bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-36_CDS_annotation_glimmer3.pl_2_1
Length=264
Score E
Sequences producing significant alignments: (Bits) Value
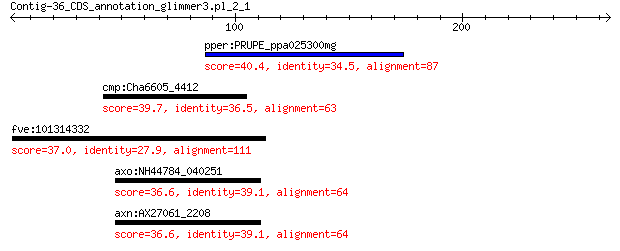
pper:PRUPE_ppa025300mg hypothetical protein 40.4 0.20
cmp:Cha6605_4412 PAS domain S-box 39.7 0.50
fve:101314332 capsid protein VP1-like 37.0 3.4
axo:NH44784_040251 Beta-carotene ketolase (EC:1.14.-.-) 36.6 5.5
axn:AX27061_2208 Beta-carotene ketolase 36.6 5.8
> pper:PRUPE_ppa025300mg hypothetical protein
Length=237
Score = 40.4 bits (93), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 30/91 (33%), Positives = 49/91 (54%), Gaps = 12/91 (13%)
Query 87 FNDR-LGRRQSYYFR-EPGYMIDMLSI-RPVY-YWATITPDYLSYRGADYFNPIYNDIGY 142
FND G+ + + +R PGY+I + +P W+ I P +RG +YFNP Y IG
Sbjct 19 FNDSSTGQSKRFLYRPRPGYIILCCGVEKPTSGSWSEIPPSTFKFRGENYFNP-YTPIGV 77
Query 143 QDISISRIANCPPQIDNVAVQEPCFNEFRAS 173
D+ + CP +I+++A Q P + +A+
Sbjct 78 -DVFV-----CPKKINHIA-QHPELPKVKAN 101
> cmp:Cha6605_4412 PAS domain S-box
Length=499
Score = 39.7 bits (91), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 23/63 (37%), Positives = 32/63 (51%), Gaps = 3/63 (5%)
Query 42 IEHVDRPKLIFSASQTVNVQVIMNQSGLNNFSGTQPLGQQGGAIAFNDRLGRRQSYYFRE 101
+EH DR IF QTVN Q N +G+ + + +GG I +LGR ++YF
Sbjct 440 LEHQDR---IFELFQTVNPQKQANSTGIGLAIVKKIVAAEGGTIRLESQLGRGTTFYFTW 496
Query 102 PGY 104
P Y
Sbjct 497 PKY 499
> fve:101314332 capsid protein VP1-like
Length=421
Score = 37.0 bits (84), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 31/115 (27%), Positives = 54/115 (47%), Gaps = 7/115 (6%)
Query 2 AGVQTIPQLAIASRLQEYKDLLGAGGSRYSDWLETFF--ASKIEHVDRPKLIFSASQTVN 59
A TI QL + ++Q+ + GG+RY++ + + F AS + RP+ + S +N
Sbjct 303 ATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPIN 362
Query 60 VQVIMNQSGLNNFSGTQPLGQQGGAIAFNDRLGRRQSY--YFREPGYMIDMLSIR 112
+ I G T P QG AF + + + F E G++I ++S+R
Sbjct 363 IAPIAQTGGTGAQGTTTP---QGNLAAFGTYMAKGHGFSQSFVEHGHVIGLVSVR 414
> axo:NH44784_040251 Beta-carotene ketolase (EC:1.14.-.-)
Length=560
Score = 36.6 bits (83), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 36/69 (52%), Gaps = 7/69 (10%)
Query 47 RPKLIFSASQTVNVQVIMNQSGLNNFSGTQPLGQQGGAIAF---NDRLGR--RQSYYFRE 101
RP+ + S T + Q+I+ SG+N+ L Q+G ++ NDRLG R F
Sbjct 14 RPEPVMPVSSTSDAQIIVVGSGMNSLVCAALLAQRGKSVLVLERNDRLGGCIRTEELF-- 71
Query 102 PGYMIDMLS 110
PGY D+LS
Sbjct 72 PGYRHDVLS 80
> axn:AX27061_2208 Beta-carotene ketolase
Length=560
Score = 36.6 bits (83), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 36/69 (52%), Gaps = 7/69 (10%)
Query 47 RPKLIFSASQTVNVQVIMNQSGLNNFSGTQPLGQQGGAIAF---NDRLGR--RQSYYFRE 101
RP+ + S T + Q+I+ SG+N+ L Q+G ++ NDRLG R F
Sbjct 14 RPEPVMPVSSTSDAQIIVVGSGMNSLVCAALLAQRGKSVLVLERNDRLGGCIRTEELF-- 71
Query 102 PGYMIDMLS 110
PGY D+LS
Sbjct 72 PGYRHDVLS 80
Lambda K H a alpha
0.319 0.134 0.393 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 402720626866