bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_2
Length=322
Score E
Sequences producing significant alignments: (Bits) Value
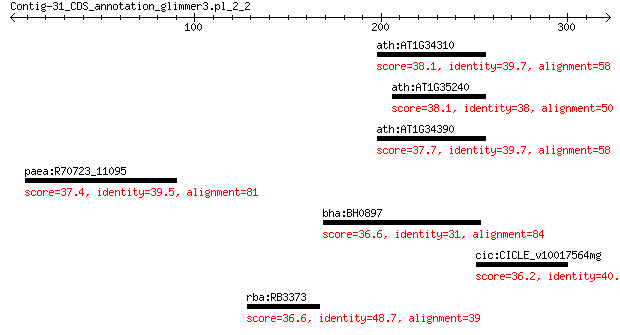
ath:AT1G34310 ARF12; auxin response factor 12 38.1
ath:AT1G35240 ARF20; auxin response factor 20 38.1
ath:AT1G34390 ARF22; auxin response factor 22 37.7
paea:R70723_11095 cystathionine beta-lyase 37.4 3.8
bha:BH0897 RNA methyltransferase 36.6 7.2
cic:CICLE_v10017564mg hypothetical protein 36.2 8.3
rba:RB3373 receptor-like histidine kinase BpdS (EC:2.7.-.-) 36.6 8.7
> ath:AT1G34310 ARF12; auxin response factor 12
Length=593
Score = 38.1 bits (87), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 23/64 (36%), Positives = 31/64 (48%), Gaps = 6/64 (9%)
Query 198 PNLDQ---IGFQELLSQEMHGRAWRVNANYKTTDFSVGKQPAWTEYTTTVNETYGD---F 251
P+LD + QELL+ ++HG WR N NY+ T W +TT+ GD F
Sbjct 153 PSLDMSQPLPAQELLAIDLHGNQWRFNHNYRGTPQRHLLTTGWNAFTTSKKLVAGDVIVF 212
Query 252 AAGE 255
GE
Sbjct 213 VRGE 216
> ath:AT1G35240 ARF20; auxin response factor 20
Length=590
Score = 38.1 bits (87), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 19/53 (36%), Positives = 27/53 (51%), Gaps = 3/53 (6%)
Query 206 QELLSQEMHGRAWRVNANYKTTDFSVGKQPAWTEYTTTVNETYGD---FAAGE 255
QELL++++HG WR +Y+ T W E+TT+ GD F GE
Sbjct 160 QELLAKDLHGNQWRFRHSYRGTPQRHSLTTGWNEFTTSKKLVKGDVIVFVRGE 212
> ath:AT1G34390 ARF22; auxin response factor 22
Length=598
Score = 37.7 bits (86), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 23/64 (36%), Positives = 30/64 (47%), Gaps = 6/64 (9%)
Query 198 PNLDQ---IGFQELLSQEMHGRAWRVNANYKTTDFSVGKQPAWTEYTTTVNETYGD---F 251
P LD + QELL+ ++HG WR N NY+ T W +TT+ GD F
Sbjct 151 PPLDMSQPLPTQELLATDLHGNQWRFNHNYRGTPQRHLLTTGWNAFTTSKKLVAGDVIVF 210
Query 252 AAGE 255
GE
Sbjct 211 VRGE 214
> paea:R70723_11095 cystathionine beta-lyase
Length=394
Score = 37.4 bits (85), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 45/96 (47%), Gaps = 17/96 (18%)
Query 9 KTESGVQVYGRTTSQCGLGIRTYLSDRFNNWLNTEWIDGTNGINEITSVDVTSGLLTM-- 66
+ ESG+ Y T I ++ S R N L TEWI + GI +TS+ + L T
Sbjct 53 RVESGIYGYSVTGESYRQAIVSWYSRRHNWELQTEWITDSPGI--VTSLSLAVELFTQPG 110
Query 67 DALILQKKV----YDMLNR---------IAVSGGSY 89
D +ILQ V YD++N + +SGG Y
Sbjct 111 DEVILQSPVYYPFYDVINMNDRKVAKNPLILSGGRY 146
> bha:BH0897 RNA methyltransferase
Length=463
Score = 36.6 bits (83), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 26/84 (31%), Positives = 39/84 (46%), Gaps = 3/84 (4%)
Query 169 ILGSIVPRVDYSQGNKWWTRVDTMNDFHKPNLDQIGFQELLSQEMHGRAWRVNANYKTTD 228
IL S+V RV + G V T DF K +L +E+ S+ H ++ + N N K T
Sbjct 203 ILRSVVSRVGFETGELQVILVTTKKDFPKKDL---LVEEIKSRLPHVKSLQQNINPKKTS 259
Query 229 FSVGKQPAWTEYTTTVNETYGDFA 252
+G + T+ ET GD +
Sbjct 260 LIMGDETISLFGEETIEETLGDIS 283
> cic:CICLE_v10017564mg hypothetical protein
Length=419
Score = 36.2 bits (82), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 20/49 (41%), Positives = 24/49 (49%), Gaps = 0/49 (0%)
Query 251 FAAGEPLEYMAFNRVYDVANDPKLKDATTYIDPQIFNKAFANSNLDAKN 299
FAAG P Y RV N PK++ P++ NK F NLD KN
Sbjct 11 FAAGLPSFYSPNARVSATMNAPKIRMMNPISLPKLSNKVFVEDNLDLKN 59
> rba:RB3373 receptor-like histidine kinase BpdS (EC:2.7.-.-)
Length=1998
Score = 36.6 bits (83), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 19/39 (49%), Positives = 21/39 (54%), Gaps = 0/39 (0%)
Query 128 AFESGETGQEPLGSLAGRGRETSKRGGKNIKIRCEEPSL 166
AFES ETG L RG KRG KN+ + C E SL
Sbjct 769 AFESAETGTHNARVLLKRGELHFKRGSKNLAVECFEASL 807
Lambda K H a alpha
0.317 0.134 0.404 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 575086215924