bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-23_CDS_annotation_glimmer3.pl_2_1
Length=221
Score E
Sequences producing significant alignments: (Bits) Value
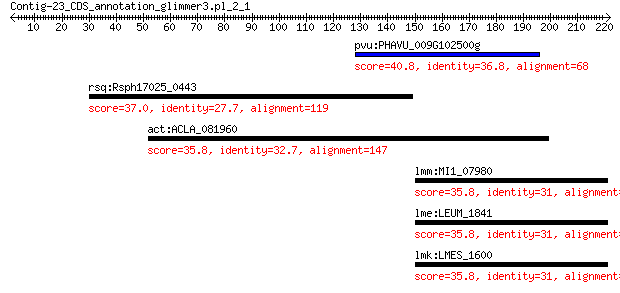
pvu:PHAVU_009G102500g hypothetical protein 40.8 0.18
rsq:Rsph17025_0443 hypothetical protein 37.0 2.8
act:ACLA_081960 C6 finger domain protein, putative 35.8 7.4
lmm:MI1_07980 septation ring formation regulator EzrA 35.8 7.5
lme:LEUM_1841 septation ring formation regulator EzrA 35.8 7.5
lmk:LMES_1600 Negative regulator of septation ring formation 35.8 7.6
> pvu:PHAVU_009G102500g hypothetical protein
Length=776
Score = 40.8 bits (94), Expect = 0.18, Method: Composition-based stats.
Identities = 25/73 (34%), Positives = 41/73 (56%), Gaps = 5/73 (7%)
Query 128 LQVQQIEQQRRMNDAQIALAEAQASKANEEAKKISGVDTQEALKRIEGIASQIEL----- 182
LQ +IE++ RM +A++A +A A E+AK + ++T EA KRI + SQ L
Sbjct 366 LQQWKIEEEMRMEEAKLAGKKAMAMAEKEKAKSKAAIETAEAQKRIAELESQKRLVAEMK 425
Query 183 NLKEGQYKEALTD 195
++E + K + D
Sbjct 426 AIRESEEKRKVLD 438
> rsq:Rsph17025_0443 hypothetical protein
Length=945
Score = 37.0 bits (84), Expect = 2.8, Method: Composition-based stats.
Identities = 33/123 (27%), Positives = 51/123 (41%), Gaps = 21/123 (17%)
Query 30 VFGLSWSPQKAMEEQWKYN-KKIMALQNQ-YQQQAAAQSQQYAKDYWDYTNAENQVEHLK 87
V G W + + W Y+ KK AL ++ Y + Q+ A WDY N E +
Sbjct 25 VMGAGWKAETIRTDAWSYSQKKRQALASEIYDRLTPDAKQRIADQRWDYENNWTDFEDMV 84
Query 88 NAGLNIGLMYGQSGAGGMGASGGAHQASPEQPQGNPVGMALQVQQIEQQRR--MNDAQIA 145
+G + A Q +PE G P+ L Q+I+ +RR M++AQ
Sbjct 85 -----------------LGEAATAAQTAPESFAGLPLSRDLFDQRIDAERRAEMDEAQAI 127
Query 146 LAE 148
L +
Sbjct 128 LDQ 130
> act:ACLA_081960 C6 finger domain protein, putative
Length=843
Score = 35.8 bits (81), Expect = 7.4, Method: Composition-based stats.
Identities = 48/177 (27%), Positives = 70/177 (40%), Gaps = 33/177 (19%)
Query 52 MALQNQYQQQAAAQSQQYAKDYWDYTNAENQVEHLKNA-------------GLNIGLMYG 98
M Y AAQ Q A Y YTN + + L +A G N G+ +G
Sbjct 9 MPPATSYPSPNAAQMGQGAMQY--YTNRQLTADELLSAELSRETSGQGLADGSNNGVHHG 66
Query 99 QSGAGGMGASGGAHQASPEQPQGNPVGMALQV---QQI-----------EQQRRMNDAQI 144
QS G GGA P P + LQ QQ+ +Q R ++I
Sbjct 67 QSMVLGSSNPGGAEMGRPSSPDQHQQQHMLQFTPSQQVGVDPNHDLSYGDQSARRKRSKI 126
Query 145 ALA--EAQASKANEEAKKISGVDTQEALKRIEGIASQI-ELNLKEGQYKEALTDLTK 198
+ A E + K +A SGV+T +R+ G+A Q + +K G K + +L +
Sbjct 127 SRACDECRRKKVRCDASSESGVETCSNCRRL-GVACQFSRVPMKRGPSKGYIKELAE 182
> lmm:MI1_07980 septation ring formation regulator EzrA
Length=571
Score = 35.8 bits (81), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 22/71 (31%), Positives = 40/71 (56%), Gaps = 2/71 (3%)
Query 150 QASKANEEAKKISGVDTQEALKRIEGIASQIELNLKEGQYKEALTDLTKAEKEATEALTS 209
QA+ EA+ I+ + T+ L+R++G+ E +K+ + L DL K ++E EA+
Sbjct 88 QANLVLFEARGINFIKTRNELERLQGMVDDTETTIKD--VRAGLLDLKKTDEEHREAIND 145
Query 210 LREMQEGLTRH 220
L++ EGL +
Sbjct 146 LKKKYEGLRKR 156
> lme:LEUM_1841 septation ring formation regulator EzrA
Length=571
Score = 35.8 bits (81), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 22/71 (31%), Positives = 40/71 (56%), Gaps = 2/71 (3%)
Query 150 QASKANEEAKKISGVDTQEALKRIEGIASQIELNLKEGQYKEALTDLTKAEKEATEALTS 209
QA+ EA+ I+ + T+ L+R++G+ E +K+ + L DL K ++E EA+
Sbjct 88 QANLVLFEARGINFIKTRNELERLQGMVDDTETTIKD--VRAGLLDLKKTDEEHREAIND 145
Query 210 LREMQEGLTRH 220
L++ EGL +
Sbjct 146 LKKKYEGLRKR 156
> lmk:LMES_1600 Negative regulator of septation ring formation
Length=571
Score = 35.8 bits (81), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 22/71 (31%), Positives = 40/71 (56%), Gaps = 2/71 (3%)
Query 150 QASKANEEAKKISGVDTQEALKRIEGIASQIELNLKEGQYKEALTDLTKAEKEATEALTS 209
QA+ EA+ I+ + T+ L+R++G+ E +K+ + L DL K ++E EA+
Sbjct 88 QANLVLFEARGINFIKTRNELERLQGMVDDTETTIKD--VRAGLLDLKKTDEEHREAIND 145
Query 210 LREMQEGLTRH 220
L++ EGL +
Sbjct 146 LKKKYEGLRKR 156
Lambda K H a alpha
0.308 0.123 0.336 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 279596245911