bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_3
Length=611
Score E
Sequences producing significant alignments: (Bits) Value
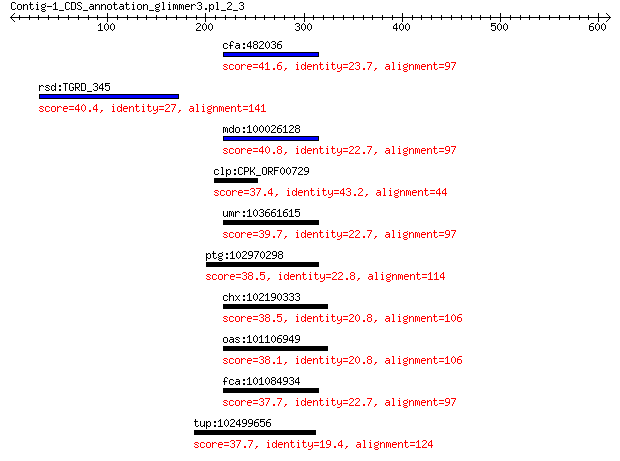
cfa:482036 FER1L6; fer-1-like 6 (C. elegans) 41.6 0.71
rsd:TGRD_345 cytoplasmic M42 family peptidase 40.4 1.1
mdo:100026128 FER1L6; fer-1-like 6 (C. elegans) 40.8 1.1
clp:CPK_ORF00729 hypothetical protein 37.4 2.1
umr:103661615 FER1L6; fer-1-like family member 6 39.7
ptg:102970298 FER1L6; fer-1-like 6 (C. elegans) 38.5 5.3
chx:102190333 FER1L6; fer-1-like 6 (C. elegans) 38.5 6.5
oas:101106949 FER1L6; fer-1-like 6 (C. elegans) 38.1 8.3
fca:101084934 FER1L6; fer-1-like 6 (C. elegans) 37.7 8.7
tup:102499656 FER1L6; fer-1-like 6 (C. elegans) 37.7 9.6
> cfa:482036 FER1L6; fer-1-like 6 (C. elegans)
Length=1858
Score = 41.6 bits (96), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 23/97 (24%), Positives = 48/97 (49%), Gaps = 8/97 (8%)
Query 218 RNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFFNSEEITQTLREDISKAWAYGRIDY 277
RN + F + K C Y +S +G QT+R HW +L E++ L E I +A +I
Sbjct 568 RNYNYLPFEAKKPCVYFISSWGDQTFRLHWSNML----EKMADLLEESIEEARELIKISE 623
Query 278 SLSRGAAASYVASYVNSAACLPFFYVGQKEIRPRSFH 314
+ S + ++ +++ ++ ++ + E +P+ +
Sbjct 624 TASEEKMKTVLSDFISQSSA----FISEAEKKPKMLN 656
> rsd:TGRD_345 cytoplasmic M42 family peptidase
Length=349
Score = 40.4 bits (93), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 38/151 (25%), Positives = 64/151 (42%), Gaps = 17/151 (11%)
Query 31 IFAPCGRCKSCIMNKSNF-ATAYAMNMATHFKYCYFVTLT-YKDIFLPYLSVEVVRRSGN 88
+ GR K IM ++ + + YF+ + D LP +VE++ + G
Sbjct 44 VIGKSGRGKKKIMIAAHMDEIGMTVKHINEKGFIYFIKIGGINDSILPGKTVEIINKKG- 102
Query 89 RYLFDENFETMVSTSDPRLLTPE-----YYHDR---DLSLDPAQNEVEQVFDIGFQSIPR 140
E+ ++ T P L+T E HD D+ L ++NEV +V DIG Q I
Sbjct 103 -----EHITGIIGTKPPHLMTSEDVKQPLKHDSMFIDIGLS-SRNEVLKVADIGDQIIFE 156
Query 141 DVSVKSKGSFRFRSFDDEPLKFCIPMKLTEL 171
V+ G+F + D + +K+ E+
Sbjct 157 PVAGILNGNFYYGKAADNRISCYTMVKVMEM 187
> mdo:100026128 FER1L6; fer-1-like 6 (C. elegans)
Length=1873
Score = 40.8 bits (94), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 22/97 (23%), Positives = 47/97 (48%), Gaps = 8/97 (8%)
Query 218 RNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFFNSEEITQTLREDISKAWAYGRIDY 277
RN F + K C Y +S +G QT+R HW +L +++ L E++ + +I
Sbjct 587 RNYHYLPFEAKKPCVYFISSWGDQTFRLHWSNML----DKMADNLEENLEEVKELIKISE 642
Query 278 SLSRGAAASYVASYVNSAACLPFFYVGQKEIRPRSFH 314
S G + + ++++ ++ ++ Q E +P+ +
Sbjct 643 EESEGKIKTALNNFISQSSA----FISQAEKKPKMLN 675
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 37.4 bits (85), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 26/44 (59%), Gaps = 5/44 (11%)
Query 209 IELFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLF 252
++LF KRLR + H KI Y+ EYG + RPH+H L+F
Sbjct 75 LQLFLKRLRDRISPH-----KIRYFGCGEYGTKLQRPHYHLLIF 113
> umr:103661615 FER1L6; fer-1-like family member 6
Length=1865
Score = 39.7 bits (91), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 22/97 (23%), Positives = 46/97 (47%), Gaps = 8/97 (8%)
Query 218 RNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFFNSEEITQTLREDISKAWAYGRIDY 277
RN + F + K C Y +S +G QT+R HW +L E++ L E I + +I
Sbjct 570 RNYNYLPFEAKKPCVYFISSWGEQTFRLHWSNML----EKMADLLEESIEEVRELIKISE 625
Query 278 SLSRGAAASYVASYVNSAACLPFFYVGQKEIRPRSFH 314
S + ++ +++ ++ ++ + E +P+ +
Sbjct 626 KASEDKMKTVLSDFISQSSA----FISEAEKKPKMLN 658
> ptg:102970298 FER1L6; fer-1-like 6 (C. elegans)
Length=1859
Score = 38.5 bits (88), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 26/115 (23%), Positives = 51/115 (44%), Gaps = 9/115 (8%)
Query 201 IPVLQSRDIE-LFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFFNSEEIT 259
IPV+ + E RN + F + K C Y +S +G QT+R HW +L E++
Sbjct 552 IPVVSTTHPEKPLVTEGNRNYNYLPFDAKKPCVYFISSWGDQTFRLHWSNML----EKMA 607
Query 260 QTLREDISKAWAYGRIDYSLSRGAAASYVASYVNSAACLPFFYVGQKEIRPRSFH 314
L E I + +I S ++ +++ ++ ++ + E +P+ +
Sbjct 608 DLLEESIEEVRELIKISEKASEDKMKMVLSDFISQSSA----FISEAEKKPKMLN 658
> chx:102190333 FER1L6; fer-1-like 6 (C. elegans)
Length=1858
Score = 38.5 bits (88), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 22/106 (21%), Positives = 47/106 (44%), Gaps = 8/106 (8%)
Query 218 RNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFFNSEEITQTLREDISKAWAYGRIDY 277
RN + F + K C Y VS +G QT+R HW +L E++ L E I + +I
Sbjct 569 RNYNYLPFEAKKPCVYFVSSWGDQTFRLHWSNML----EKMADLLEEGIEEVKELIKISE 624
Query 278 SLSRGAAASYVASYVNSAACLPFFYVGQKEIRPRSFHSKGFGSNKI 323
++ +++ ++ ++ + E++P+ + ++
Sbjct 625 EAPEEKMKLVLSDFISQSSA----FISEAEMKPKMLNQTNLDKKRL 666
> oas:101106949 FER1L6; fer-1-like 6 (C. elegans)
Length=1859
Score = 38.1 bits (87), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 22/106 (21%), Positives = 47/106 (44%), Gaps = 8/106 (8%)
Query 218 RNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFFNSEEITQTLREDISKAWAYGRIDY 277
RN + F + K C Y VS +G QT+R HW +L E++ L E I + +I
Sbjct 570 RNYNYLPFEAKKPCVYFVSSWGDQTFRLHWSNML----EKMADFLEEGIEEVKELIKISE 625
Query 278 SLSRGAAASYVASYVNSAACLPFFYVGQKEIRPRSFHSKGFGSNKI 323
++ +++ ++ ++ + E++P+ + ++
Sbjct 626 EAPEEKMKMVLSDFISQSSA----FISEAEMKPKMLNQTNLDKKRL 667
> fca:101084934 FER1L6; fer-1-like 6 (C. elegans)
Length=1859
Score = 37.7 bits (86), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 22/97 (23%), Positives = 45/97 (46%), Gaps = 8/97 (8%)
Query 218 RNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFFNSEEITQTLREDISKAWAYGRIDY 277
RN + F + K C Y +S +G QT+R HW +L E++ L E I + +I
Sbjct 570 RNYNYLPFDAKKPCVYFISSWGDQTFRLHWSNML----EKMADLLEESIEEVRELIKISE 625
Query 278 SLSRGAAASYVASYVNSAACLPFFYVGQKEIRPRSFH 314
S ++ +++ ++ ++ + E +P+ +
Sbjct 626 KASEDKMKMVLSDFISQSSA----FISEAEKKPKMLN 658
> tup:102499656 FER1L6; fer-1-like 6 (C. elegans)
Length=1858
Score = 37.7 bits (86), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 24/124 (19%), Positives = 56/124 (45%), Gaps = 17/124 (14%)
Query 188 KVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTYRPHW 247
+V +P ++ + + P++ + RN + F + K C Y +S +G QT+R HW
Sbjct 547 EVAFPMVSTTQPEKPLVTEGN---------RNYNYLPFEAKKPCVYFISSWGDQTFRLHW 597
Query 248 HCLLFFNSEEITQTLREDISKAWAYGRIDYSLSRGAAASYVASYVNSAACLPFFYVGQKE 307
+L E++ L E I + +I + ++ +++ ++ ++ + E
Sbjct 598 SNVL----EKMADLLEESIEEVRELIKISEDAPEEKMKTVLSDFISQSSA----FIAEAE 649
Query 308 IRPR 311
+P+
Sbjct 650 KKPK 653
Lambda K H a alpha
0.324 0.139 0.434 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1414931933445