bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-19_CDS_annotation_glimmer3.pl_2_3
Length=639
Score E
Sequences producing significant alignments: (Bits) Value
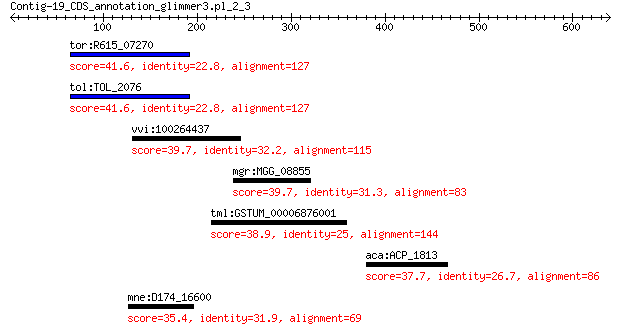
tor:R615_07270 Na(+)-translocating NADH-quinone reductase subu... 41.6 0.54
tol:TOL_2076 Na(+)-translocating NADH-quinone reductase subunit A 41.6 0.54
vvi:100264437 methionine S-methyltransferase-like 39.7 2.2
mgr:MGG_08855 hypothetical protein 39.7 2.6
tml:GSTUM_00006876001 hypothetical protein 38.9 4.0
aca:ACP_1813 ImpB/MucB/SamB family protein 37.7 8.7
mne:D174_16600 hypothetical protein 35.4 9.5
> tor:R615_07270 Na(+)-translocating NADH-quinone reductase subunit
A
Length=444
Score = 41.6 bits (96), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 29/133 (22%), Positives = 57/133 (43%), Gaps = 9/133 (7%)
Query 65 LMNGYKLITIATFTPDSAIYGWMRNGRRYEPDEYTKFGKVYFPLAGSNIGNYIDPAFQIA 124
+ +G K+ +A PD Y M+ + + K G+V F + Y PA +
Sbjct 20 ITDGPKVTQVAVMGPD---YVGMKPTMAVQEGDRVKKGQVLFTDKKTEGVQYTAPAAGVV 76
Query 125 RRVRR------LTFGLDLDANRAKIYESWVSDELNGTGDNGQPTHIGRGGLWDWLGIAPG 178
+ + R L+ ++LD + + + S+ ++LN ++ GLW L P
Sbjct 77 KAINRGARRVFLSVVIELDGDEEETFTSYTPEQLNSLDAKDVEANLVASGLWTALRTRPY 136
Query 179 AVCPNLGAKLSSV 191
+ P LG++ ++
Sbjct 137 SKVPALGSRPKAI 149
> tol:TOL_2076 Na(+)-translocating NADH-quinone reductase subunit
A
Length=444
Score = 41.6 bits (96), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 29/133 (22%), Positives = 57/133 (43%), Gaps = 9/133 (7%)
Query 65 LMNGYKLITIATFTPDSAIYGWMRNGRRYEPDEYTKFGKVYFPLAGSNIGNYIDPAFQIA 124
+ +G K+ +A PD Y M+ + + K G+V F + Y PA +
Sbjct 20 ITDGPKVTQVAVMGPD---YVGMKPTMAVQEGDRVKKGQVLFTDKKTEGVQYTAPAAGVV 76
Query 125 RRVRR------LTFGLDLDANRAKIYESWVSDELNGTGDNGQPTHIGRGGLWDWLGIAPG 178
+ + R L+ ++LD + + + S+ ++LN ++ GLW L P
Sbjct 77 KAINRGARRVFLSVVIELDGDEEETFTSYTPEQLNSLDAKDVEANLVASGLWTALRTRPY 136
Query 179 AVCPNLGAKLSSV 191
+ P LG++ ++
Sbjct 137 SKVPALGSRPKAI 149
> vvi:100264437 methionine S-methyltransferase-like
Length=1092
Score = 39.7 bits (91), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 56/130 (43%), Gaps = 17/130 (13%)
Query 131 TFGLDLDANRAKIYESWVSDELNGTGDNGQPTHIGRGG-LWDWLGIAPG---AVCPNLGA 186
+GLD++ KI SW++ LN DNGQP + G L D + A C + G
Sbjct 145 VYGLDINPRAVKI--SWINLYLNALDDNGQPIYDGENKTLLDRVEFHESDLLAYCRDRGI 202
Query 187 KLSSVAGIRGQLYPPS-------FKFNAAPFFAYFLSHYYYIANMQENYM---YFTRGVG 236
+L + G Q+ P+ NA+ F Y LS+Y + E+ R V
Sbjct 203 ELERIVGCIPQILNPNPDAMSKMITENASEEFLYSLSNYCALQGFVEDQFGLGLIARAVE 262
Query 237 E-MQKVRPDG 245
E + ++P G
Sbjct 263 EGIAVIKPMG 272
> mgr:MGG_08855 hypothetical protein
Length=1020
Score = 39.7 bits (91), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 26/85 (31%), Positives = 38/85 (45%), Gaps = 2/85 (2%)
Query 238 MQKVRPDGQQESLYRPFFSDVFTSFKPNEFLLYLDTLAYRTREGAQINAMDLKVEGLPIN 297
M VR G E L FFSD F+ +PN DT+ RT + + + V L +
Sbjct 290 MSTVRSSGSHEGLSDAFFSDAFSDDRPNSPASTADTIPERTHDASPPSPPATAVAQLDDD 349
Query 298 CV--QAMACAGIQGYGGLLSVPYSP 320
+ Q+ + + GGL VP +P
Sbjct 350 LIGPQSSSPWNYRMVGGLRKVPSTP 374
> tml:GSTUM_00006876001 hypothetical protein
Length=574
Score = 38.9 bits (89), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 36/145 (25%), Positives = 61/145 (42%), Gaps = 11/145 (8%)
Query 215 LSHYYYIANMQENYMYFTRGVGEMQKVRPDGQQESLYRPFFSDVFTSFKPNEFLL-YLDT 273
LS +Y +N + +Y ++ V + + + +R DV + P E +L +DT
Sbjct 157 LSQLFYYSNNHRDGVY-----SKLAPVEINDKPKFQHRGLNMDVARQWYPKEEILKIIDT 211
Query 274 LAYRTREGAQINAMDLKVEGLPINCVQAMACAGIQGYGGLLSVPYSPDLFSNIIKQGSSP 333
L++ ++ D + L I + +A G G + YSP +I+ G S
Sbjct 212 LSWNKMNRLHLHVTDSQSWPLEIPAMPNLAARGAYADG----LTYSPQDLQDILTWGRSR 267
Query 334 SIEITVENDQPNKPDAGFYVAVPEL 358
+E+ VE D P A PEL
Sbjct 268 GVEVIVEIDMPGH-TTSIAEAYPEL 291
> aca:ACP_1813 ImpB/MucB/SamB family protein
Length=441
Score = 37.7 bits (86), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 23/86 (27%), Positives = 39/86 (45%), Gaps = 5/86 (6%)
Query 380 DVFRTLWGAKSSAPYVNKPDFLGVWQASVNPSNVRAMANGSASGEDANLGQLAACIDRYC 439
+ R LW + + P KP F+GVW ++ P ++ ++ S+ +A L IDR
Sbjct 349 EALRKLWELRPTGPAHQKPFFVGVWLGNLVPDHLHTLSLFSSLETEARRTSLTTTIDR-- 406
Query 440 DFSGHSGIDYYAKEPGTFMLIAMLVP 465
+ G+D A P + +L P
Sbjct 407 -VNLKYGLDTLA--PASMLLAKAAAP 429
> mne:D174_16600 hypothetical protein
Length=114
Score = 35.4 bits (80), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 22/69 (32%), Positives = 36/69 (52%), Gaps = 11/69 (16%)
Query 127 VRRLTFGLDLDANRAKIYESWVSDELNGTGDNGQPTHIGRGGLWDWLGIAPGAVCPNLGA 186
RRL+ G+D DA+++ ++ +S L T + GQ +W +LG A CP A
Sbjct 56 CRRLSTGVDADASQSAVF---LSRNLARTMNTGQ--------VWQFLGTAVSLYCPQHTA 104
Query 187 KLSSVAGIR 195
L+ ++G R
Sbjct 105 ALAEMSGDR 113
Lambda K H a alpha
0.321 0.139 0.439 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1500132135889