bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-14_CDS_annotation_glimmer3.pl_2_4
Length=596
Score E
Sequences producing significant alignments: (Bits) Value
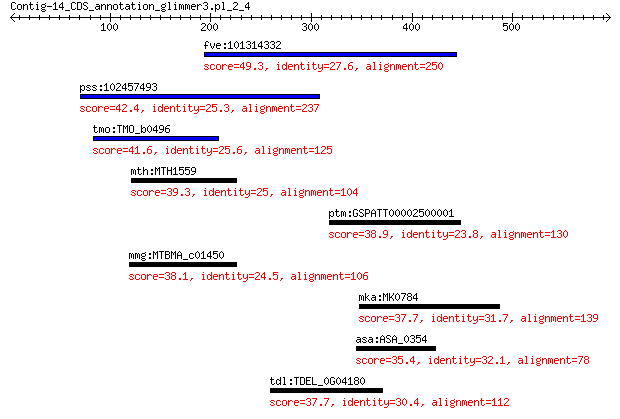
fve:101314332 capsid protein VP1-like 49.3 0.002
pss:102457493 DGCR14; DiGeorge syndrome critical region gene 14 42.4
tmo:TMO_b0496 caiC; AMP-dependent synthetase and ligase 41.6 0.51
mth:MTH1559 tungsten formylmethanofuran dehydrogenase subunit B 39.3 2.3
ptm:GSPATT00002500001 hypothetical protein 38.9 3.9
mmg:MTBMA_c01450 fwdB; tungsten formylmethanofuran dehydrogena... 38.1 5.2
mka:MK0784 TrpB; tryptophan synthase subunit beta (EC:4.2.1.20) 37.7 6.6
asa:ASA_0354 lfiE; lateral flagellar hook basal body protein, ... 35.4 6.6
tdl:TDEL_0G04180 TDEL0G04180; hypothetical protein 37.7 7.6
> fve:101314332 capsid protein VP1-like
Length=421
Score = 49.3 bits (116), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 69/264 (26%), Positives = 105/264 (40%), Gaps = 33/264 (13%)
Query 194 NVLPLAAYQKIYCDYFRFEQWEKAQPYTYNFDYYSGGNILTEYKGDPSDLFLKDNLFSLR 253
+ LP+ AY IY +FR E + N + KGD D N LR
Sbjct 176 SALPVRAYNLIYNQWFRDENLQ---------------NSVVVDKGDGPDTTPSTNYTLLR 220
Query 254 YANYPKDLFMGILPSSQLGSVATINISHSSSAGVHLTNQEGYLTGTVAS-DGTTI----T 308
D F LP Q G A +++ +SA + + G G +++ G I +
Sbjct 221 RGKR-HDYFTSALPWPQKGGTA-VSLPLGTSAPIAFSGASGSDVGVISTTQGNLIKNMYS 278
Query 309 VKNTRSLTPGISPVLRTNFADLNAN--FDILSFRIANAIQRMREIQQCAGQGYKEQLEAR 366
+ SL G + V +ADL+A I R + IQ++ E G Y E + +
Sbjct 279 TGSGTSLKIGSATVATGLYADLSAATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSH 338
Query 367 WNVKLSTALSDHCTYIGGNSSQINISEVLNNSLDTEQS----QADIKGKG--VGSGSG-S 419
+ V A Y+GG S+ INI+ + Q Q ++ G + G G S
Sbjct 339 FGVASPDARLQRPEYLGGGSTPINIAPIAQTGGTGAQGTTTPQGNLAAFGTYMAKGHGFS 398
Query 420 ESFETQEHGILMCIYHAVPVLDYQ 443
+SF EHG ++ + L YQ
Sbjct 399 QSFV--EHGHVIGLVSVRADLTYQ 420
> pss:102457493 DGCR14; DiGeorge syndrome critical region gene
14
Length=399
Score = 42.4 bits (98), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 60/238 (25%), Positives = 86/238 (36%), Gaps = 47/238 (20%)
Query 71 KEYFDWYFVPLRLINKNLNPALVNMLDQSNIANSLIQNKVVSDEIPCFDYDTLTSCLKAF 130
KE Y P ++P ++L++S I + V E + D L S L +F
Sbjct 17 KETPAPYVTPATFETPEVHPGASSLLNKSKIGIKTAEEGEVEKEQ---NKDALPS-LDSF 72
Query 131 NTQHPSYLDIAGFERVPKTLKLLRYLRYGNFLYDTGFSTLPSKNMNYSSVKDFNLYAKWN 190
T+H S D A FE++ + K R+ ++LY+ +A+
Sbjct 73 LTKHTSE-DNASFEQIMEVAKEKEKARH-SWLYEAE-----------------EEFAQRR 113
Query 191 LNVNVLPLAAYQKIYCDYFRFEQWE-KAQPYTYNFDYYSGGNILTEYKGDPSDLFLKDNL 249
L LP A +Q + E WE KAQ + YY G D D+F K
Sbjct 114 LEYLALPSAEHQALESGKAGVETWEYKAQ---NSLMYYPPGVP------DEDDVFKKPRE 164
Query 250 FSLRYANYPKDLFMGILPSSQLGSVATINISHSSSAGVHLTNQEGYLTGTVASDGTTI 307
R + KD F L SQL A +N Y G V DG +
Sbjct 165 VVHRNTRFLKDPFSQALSKSQLQQAAALNAQ--------------YKQGKVGPDGKEL 208
> tmo:TMO_b0496 caiC; AMP-dependent synthetase and ligase
Length=584
Score = 41.6 bits (96), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 32/127 (25%), Positives = 57/127 (45%), Gaps = 12/127 (9%)
Query 83 LINKNLNPALVNMLDQSNIANSLIQNKVVSDEIPCFDYDTLTSCLKAFNTQHPSYLDIAG 142
+ + +L PA+ +LD+ ++AN ++ + SD D+D + L P D+A
Sbjct 119 ITDADLAPAVAAVLDEVSLANIVVHRPLPSDH----DHDPAKAALPPALLARPGLHDLAE 174
Query 143 FERVPKTLKLLRY--LRYGNFLYDTGFSTLPSKNMNYSSVKDFNLYAKWNLNVNVLPLAA 200
R P+ + + N +Y +G +T P+K + V+ F ++ N L A
Sbjct 175 LMRTPRRHRPVTRGAADIANIIYTSG-TTGPAKGV----VQPFRWINQYTFNSRAF-LTA 228
Query 201 YQKIYCD 207
IYCD
Sbjct 229 KDTIYCD 235
> mth:MTH1559 tungsten formylmethanofuran dehydrogenase subunit
B
Length=437
Score = 39.3 bits (90), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 26/110 (24%), Positives = 51/110 (46%), Gaps = 14/110 (13%)
Query 121 DTLTSCLKAFNTQHPSYLDIAGF--ERVPKTLKLLRYLRYGNFLYDTGFSTLPSKNMNYS 178
D + SCL Y ++AG E++ + +++L+ ++G + G + K+ N
Sbjct 216 DAMRSCLLGHEIL---YDEVAGVPREQIEEAVEVLKNAQFGILFFGMGITHSRGKHRNID 272
Query 179 S----VKDFNLYAKWNLNVNVLPLAAYQKIYCDYFRFEQWEKAQPYTYNF 224
+ V+D N YAKW L +P+ + + + + WE P+ +F
Sbjct 273 TAIMMVQDLNDYAKWTL----IPMRGHYNVT-GFNQVCTWESGYPFCVDF 317
> ptm:GSPATT00002500001 hypothetical protein
Length=773
Score = 38.9 bits (89), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 31/139 (22%), Positives = 60/139 (43%), Gaps = 12/139 (9%)
Query 318 GISPVLRTNFADLNANFDILSFRIANAIQRMREIQQCAGQGYKEQLEARWNVKL-STALS 376
G+S +R NF + + R + +++ + QC E+L +WN++L S L
Sbjct 279 GLSNEMRNNFQTMKQLSTVTKPRGVDRVKQADQFIQCFHNKESEELIKKWNIQLESKCLQ 338
Query 377 DHCT------YIGGNSSQINISEVLNNSLDTEQSQADIKGKGVGSGSG--SESFETQEHG 428
+ I GN++QINI +LD + A ++G G+ + + + Q
Sbjct 339 IQSSKVKPGNIIMGNNTQINIE---TGNLDRDTQTAMLRGVGLENWAILYGDRDSRQAED 395
Query 429 ILMCIYHAVPVLDYQLTGP 447
+ C+ ++ +Q P
Sbjct 396 FMSCLRESIEYCKFQCKAP 414
> mmg:MTBMA_c01450 fwdB; tungsten formylmethanofuran dehydrogenase,
subunit B (EC:1.2.99.5)
Length=432
Score = 38.1 bits (87), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 26/112 (23%), Positives = 51/112 (46%), Gaps = 11/112 (10%)
Query 119 DYDTLTSCLKAFNTQHPSYLDIAGF--ERVPKTLKLLRYLRYGNFLYDTGFSTLPSKNMN 176
DY+ L + Y ++AG E++ + +++L+ ++G + G + K+ N
Sbjct 206 DYELLDAMRAYLLGNEILYDEVAGVPREQIEEAVEVLKKAQFGILFFGMGITHSRGKHRN 265
Query 177 YSS----VKDFNLYAKWNLNVNVLPLAAYQKIYCDYFRFEQWEKAQPYTYNF 224
+ V+D N YAKW L +P+ + + + + WE PY +F
Sbjct 266 IDTAIMMVEDLNDYAKWTL----IPMRGHYNV-TGFNQVCTWESGYPYCVDF 312
> mka:MK0784 TrpB; tryptophan synthase subunit beta (EC:4.2.1.20)
Length=398
Score = 37.7 bits (86), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 44/157 (28%), Positives = 59/157 (38%), Gaps = 28/157 (18%)
Query 348 MREIQQCAGQGYKEQLEARWNVKLSTALSDH---CTYIGGNSSQINI------------- 391
+RE Q+ G+ KEQ+ +L L D CT GG S+ I I
Sbjct 203 VREFQRVIGRETKEQI-----TELEGGLPDAIVACT--GGGSNSIGIFYDFLDDEEVALY 255
Query 392 -SEVLNNSLDTEQSQADIKGKGVGSGSGSESFETQ-EHGILMCIYHAVPVLDYQLTGPDL 449
E LDT++ A + VG G + Q EHG + + P LDY GP+L
Sbjct 256 AVEAGGKGLDTDEHSASLCAGEVGVLHGCRTKVLQDEHGQIRPTHSIAPGLDYPGVGPEL 315
Query 450 QLLNTYATDLPQPELDNLGLEALPYFTFVNDAVATQP 486
L D EAL F +N+ P
Sbjct 316 AFLVDEGRVTADAVTDE---EALRGFVMLNETEGILP 349
> asa:ASA_0354 lfiE; lateral flagellar hook basal body protein,
LfiE
Length=108
Score = 35.4 bits (80), Expect = 6.6, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 39/78 (50%), Gaps = 4/78 (5%)
Query 345 IQRMREIQQCAGQGYKEQLEARWNVKLSTALSDHCTYIGGNSSQINISEVLNNSLDTEQS 404
+Q+M+E+Q+ A Q +L NV + A+ D +G + Q NI+ L ++D QS
Sbjct 13 LQQMQEMQRVANQAPSIELNDASNVSFTDAMRD---VVGRVNEQQNIASKLMTAVDAGQS 69
Query 405 QADIKGKGVGSGSGSESF 422
D+ G V S SF
Sbjct 70 D-DLVGAMVASQKAGLSF 86
> tdl:TDEL_0G04180 TDEL0G04180; hypothetical protein
Length=821
Score = 37.7 bits (86), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 52/112 (46%), Gaps = 13/112 (12%)
Query 259 KDLFMGILPSSQLGSVATINISHSSSAGVHLTNQEGYLTGTVASDGTTITVKNTRSLTPG 318
KDLF LP+ + G+ A IN++H S +H + Q G L V+S TI KN L
Sbjct 9 KDLFGVALPNLECGTGAAINVNH-CSMDLHHSRQFGTLLKIVSSRDHTI--KNLEVLQAV 65
Query 319 ISPVLRTNFADLNANFDILSFRIANAIQRMREIQQCAGQGYKEQLEARWNVK 370
I R + A L +N +F + + R++Q C +L +W +
Sbjct 66 IQVGTRLSKAKLISN----NFWWTLMVNKCRDVQWCV------ELSPKWRTR 107
Lambda K H a alpha
0.319 0.136 0.408 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1369288967850