bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-10_CDS_annotation_glimmer3.pl_2_4
Length=67
Score E
Sequences producing significant alignments: (Bits) Value
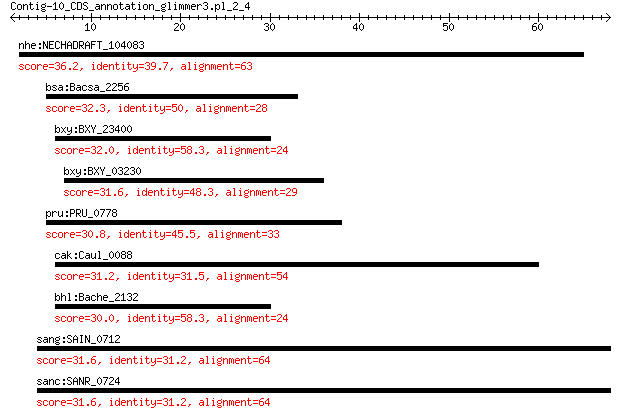
nhe:NECHADRAFT_104083 hypothetical protein 36.2 0.38
bsa:Bacsa_2256 hypothetical protein 32.3 1.1
bxy:BXY_23400 hypothetical protein 32.0 1.3
bxy:BXY_03230 hypothetical protein 31.6 2.1
pru:PRU_0778 lipoprotein 30.8 3.1
cak:Caul_0088 hypothetical protein 31.2 3.8
bhl:Bache_2132 hypothetical protein 30.0 6.8
sang:SAIN_0712 hypothetical protein 31.6 9.2
sanc:SANR_0724 hypothetical protein 31.6 9.7
> nhe:NECHADRAFT_104083 hypothetical protein
Length=598
Score = 36.2 bits (82), Expect = 0.38, Method: Composition-based stats.
Identities = 25/64 (39%), Positives = 32/64 (50%), Gaps = 4/64 (6%)
Query 2 VTKTKKEIWILILKIVAAIAAAILGV-LGVQSCTMSMSIAKNNNNATQQTEQKATSEVKN 60
VTK K++ WILIL IVAA IL + LG+ + + + Q Q TS N
Sbjct 19 VTKPKRKRWILILSIVAAAIVVILALTLGLY---FGLKKSSGGGGGSNQESQDGTSTDSN 75
Query 61 DSIN 64
DS N
Sbjct 76 DSTN 79
> bsa:Bacsa_2256 hypothetical protein
Length=31
Score = 32.3 bits (72), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 5 TKKEIWILILKIVAAIAAAILGVLGVQS 32
TKK +W L+LK++ A+A+AI G LG +
Sbjct 2 TKKGVWDLVLKVIIAVASAIAGALGANA 29
> bxy:BXY_23400 hypothetical protein
Length=34
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/24 (58%), Positives = 20/24 (83%), Gaps = 0/24 (0%)
Query 6 KKEIWILILKIVAAIAAAILGVLG 29
KK +W +ILK+V A+A+A+ GVLG
Sbjct 5 KKSVWDMILKVVIAVASALAGVLG 28
> bxy:BXY_03230 hypothetical protein
Length=32
Score = 31.6 bits (70), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 22/29 (76%), Gaps = 0/29 (0%)
Query 7 KEIWILILKIVAAIAAAILGVLGVQSCTM 35
K+ W +ILK++ A+A AI GV+GVQ+ +
Sbjct 4 KKTWSIILKVIIAVAGAIAGVVGVQAANL 32
> pru:PRU_0778 lipoprotein
Length=34
Score = 30.8 bits (68), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 23/33 (70%), Gaps = 0/33 (0%)
Query 5 TKKEIWILILKIVAAIAAAILGVLGVQSCTMSM 37
TKKE + I++++A+IA AI+ LG SC +M
Sbjct 2 TKKETFKFIIQMIASIATAIVTALGATSCMGAM 34
> cak:Caul_0088 hypothetical protein
Length=83
Score = 31.2 bits (69), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 28/54 (52%), Gaps = 1/54 (2%)
Query 6 KKEIWILILKIVAAIAAAILGVLGVQSCTMSMSIAKNNNNATQQTEQKATSEVK 59
K W+LI+ V A AAA+ G G++S ++ N TQ+ E+ +T +
Sbjct 24 KHVFWVLIISTVLA-AAALFGAWGLRSGDLAGVEHNNGAKTTQEAERYSTEQAP 76
> bhl:Bache_2132 hypothetical protein
Length=31
Score = 30.0 bits (66), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 14/24 (58%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 6 KKEIWILILKIVAAIAAAILGVLG 29
KK +W ILK+V A+A+AI+G LG
Sbjct 2 KKSVWDKILKVVIAVASAIIGALG 25
> sang:SAIN_0712 hypothetical protein
Length=242
Score = 31.6 bits (70), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 35/64 (55%), Gaps = 9/64 (14%)
Query 4 KTKKEIWILILKIVAAIAAAILGVLGVQSCTMSMSIAKNNNNATQQTEQKATSEVKNDSI 63
K +K+IW +I I A GVL + + T + +NN+NA Q T + T+ + + I
Sbjct 2 KNRKQIWYIISSIFFA------GVLFIYATTTNY---QNNSNARQTTSENYTNTLPSVPI 52
Query 64 NLKF 67
++K+
Sbjct 53 DIKY 56
> sanc:SANR_0724 hypothetical protein
Length=242
Score = 31.6 bits (70), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 35/64 (55%), Gaps = 9/64 (14%)
Query 4 KTKKEIWILILKIVAAIAAAILGVLGVQSCTMSMSIAKNNNNATQQTEQKATSEVKNDSI 63
K +K+IW +I I A GVL + + T + +NN+NA Q T + T+ + + I
Sbjct 2 KNRKQIWYIISSIFFA------GVLFIYATTTNY---QNNSNARQTTSETYTNTLPSVPI 52
Query 64 NLKF 67
++K+
Sbjct 53 DIKY 56
Lambda K H a alpha
0.313 0.123 0.320 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 127864635584