bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-6_CDS_annotation_glimmer3.pl_2_6
Length=94
Score E
Sequences producing significant alignments: (Bits) Value
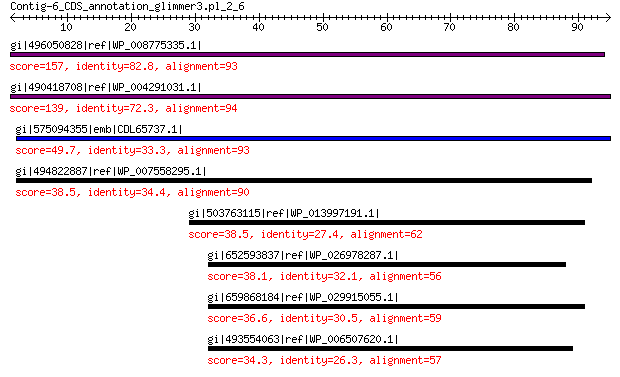
gi|496050828|ref|WP_008775335.1| hypothetical protein 157 1e-43
gi|490418708|ref|WP_004291031.1| hypothetical protein 139 6e-37
gi|575094355|emb|CDL65737.1| unnamed protein product 49.7 7e-05
gi|494822887|ref|WP_007558295.1| hypothetical protein 38.5 0.36
gi|503763115|ref|WP_013997191.1| ATP-dependent DNA helicase RecQ 38.5 0.38
gi|652593837|ref|WP_026978287.1| ATP-dependent DNA helicase RecQ 38.1 0.43
gi|659868184|ref|WP_029915055.1| ATP-dependent DNA helicase RecQ 36.6 1.6
gi|493554063|ref|WP_006507620.1| ATP-dependent DNA helicase RecQ 34.3 7.4
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 157 bits (398), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 77/94 (82%), Positives = 85/94 (90%), Gaps = 1/94 (1%)
Query 1 MIEEFYSYLDYMHLTTFFESQQEFYESDLIGDDDLCTDQWENSYYPYFYYNVHTNEP-FE 59
+IEEFYS LDYMHLT FFE+QQ FYESDLIGDDDLCTD W+NSYYPYFY NV+T+ FE
Sbjct 404 LIEEFYSRLDYMHLTKFFEAQQLFYESDLIGDDDLCTDNWDNSYYPYFYNNVYTDTNLFE 463
Query 60 KTPVYRLYASDVKKLFNDRIKHKKLNDANKIFID 93
KTPVYRLY+SDVKKLFNDRIKHKKLNDANK+F +
Sbjct 464 KTPVYRLYSSDVKKLFNDRIKHKKLNDANKVFFE 497
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 139 bits (349), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 68/95 (72%), Positives = 81/95 (85%), Gaps = 1/95 (1%)
Query 1 MIEEFYSYLDYMHLTTFFESQQEFYESDLIGDDDLCTDQWENSYYPYFYYNVH-TNEPFE 59
+IEEFYS LDYMHL TFFE+QQ FYESDL+GD DL +D WENSYYP+FY NV+ ++E ++
Sbjct 328 LIEEFYSRLDYMHLKTFFENQQLFYESDLVGDLDLMSDAWENSYYPFFYDNVYFSSEVYK 387
Query 60 KTPVYRLYASDVKKLFNDRIKHKKLNDANKIFIDE 94
KTPVYRLY + KLF+DRIKHKKLND NKIF+DE
Sbjct 388 KTPVYRLYDMQISKLFSDRIKHKKLNDLNKIFVDE 422
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 49.7 bits (117), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 31/94 (33%), Positives = 51/94 (54%), Gaps = 11/94 (12%)
Query 2 IEEFYSYLDYMHLTTFFESQQEFYESDLIGDDDLCTDQWENSYYPYFYYNVH-TNEPFEK 60
IE FY LDY+ LT FF+SQ+ ++ D DD Y Y Y N + + +++
Sbjct 434 IEAFYKDLDYLQLTEFFKSQELYFSQDFCDSDD----------YVYMYNNSSFSLDAYKQ 483
Query 61 TPVYRLYASDVKKLFNDRIKHKKLNDANKIFIDE 94
+ Y + +++ +IKHK+LND N++F D+
Sbjct 484 SMSYLSFEQQTFEIWRSKIKHKELNDLNQLFFDK 517
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 38.5 bits (88), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 31/99 (31%), Positives = 54/99 (55%), Gaps = 15/99 (15%)
Query 2 IEEFYSYLDYMHLTTFFESQQEFY------ESDLI-GDDDLCTDQWENSYYPYFYYNV-H 53
+E F+ +Y++L ++ Q+ ++ +SD + G + L D YFY NV +
Sbjct 449 VERFWKNYEYLNLVDWYRKQEIYFDKSYSRKSDFLDGKERLSGD------IKYFYNNVPY 502
Query 54 TNEPFEK-TPVYRLYASDVKKLFNDRIKHKKLNDANKIF 91
E F+K T Y+ Y+++V+ + R+KHK+ ND N IF
Sbjct 503 DVEQFKKRTFAYKAYSANVRFMARQRMKHKEQNDKNMIF 541
>gi|503763115|ref|WP_013997191.1| ATP-dependent DNA helicase RecQ [Capnocytophaga canimorsus]
gi|340621856|ref|YP_004740308.1| ATP-dependent DNA helicase recQ [Capnocytophaga canimorsus Cc5]
gi|339902122|gb|AEK23201.1| ATP-dependent DNA helicase recQ [Capnocytophaga canimorsus Cc5]
Length=635
Score = 38.5 bits (88), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 30/62 (48%), Gaps = 0/62 (0%)
Query 29 LIGDDDLCTDQWENSYYPYFYYNVHTNEPFEKTPVYRLYASDVKKLFNDRIKHKKLNDAN 88
++ D+ C W + P + V E F K P+ L AS +++ ND ++ ++ND
Sbjct 135 IVIDEAHCISHWGKDFRPAYLECVWLKEEFPKIPILALTASATQRVQNDIVQLMQMNDVQ 194
Query 89 KI 90
I
Sbjct 195 VI 196
>gi|652593837|ref|WP_026978287.1| ATP-dependent DNA helicase RecQ [Flavobacterium tegetincola]
Length=631
Score = 38.1 bits (87), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 32 DDDLCTDQWENSYYPYFYYNVHTNEPFEKTPVYRLYASDVKKLFNDRIKHKKLNDA 87
D+ C QW N + P + + E F KTP+ L AS ++ D ++ K+ DA
Sbjct 137 DEAHCISQWGNDFRPAYLKITNLKEHFPKTPILALTASATPRVKLDILRILKIEDA 192
>gi|659868184|ref|WP_029915055.1| ATP-dependent DNA helicase RecQ [Pelobacter seleniigenes]
Length=594
Score = 36.6 bits (83), Expect = 1.6, Method: Composition-based stats.
Identities = 18/59 (31%), Positives = 29/59 (49%), Gaps = 0/59 (0%)
Query 32 DDDLCTDQWENSYYPYFYYNVHTNEPFEKTPVYRLYASDVKKLFNDRIKHKKLNDANKI 90
D+ C QW + + P + E F P+ L A+ K+ ND I+H L DA+++
Sbjct 134 DEAHCVSQWGHDFRPEYVELGRLREEFPDVPMIALTATAEKQTRNDIIRHLCLQDAHQV 192
>gi|493554063|ref|WP_006507620.1| ATP-dependent DNA helicase RecQ [Xenococcus sp. PCC 7305]
gi|442795958|gb|ELS05292.1| ATP-dependent DNA helicase RecQ [Xenococcus sp. PCC 7305]
Length=709
Score = 34.3 bits (77), Expect = 7.4, Method: Composition-based stats.
Identities = 15/57 (26%), Positives = 28/57 (49%), Gaps = 0/57 (0%)
Query 32 DDDLCTDQWENSYYPYFYYNVHTNEPFEKTPVYRLYASDVKKLFNDRIKHKKLNDAN 88
D+ C +W + + P + F +TP++ A+ K++ +D I+ L DAN
Sbjct 142 DEAHCVSEWGHDFRPEYRQLKQLRSQFPQTPIFAFTATATKRVQDDIIQQLGLRDAN 198
Lambda K H a alpha
0.320 0.139 0.428 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 432797507010