bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-6_CDS_annotation_glimmer3.pl_2_3
Length=357
Score E
Sequences producing significant alignments: (Bits) Value
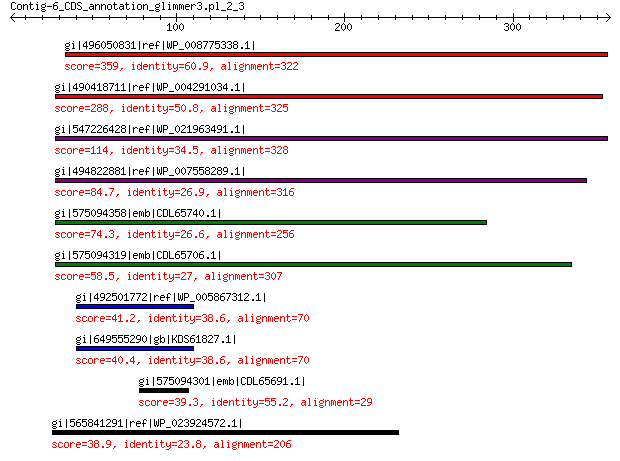
gi|496050831|ref|WP_008775338.1| predicted protein 359 4e-118
gi|490418711|ref|WP_004291034.1| hypothetical protein 288 1e-90
gi|547226428|ref|WP_021963491.1| putative uncharacterized protein 114 3e-25
gi|494822881|ref|WP_007558289.1| hypothetical protein 84.7 3e-15
gi|575094358|emb|CDL65740.1| unnamed protein product 74.3 1e-11
gi|575094319|emb|CDL65706.1| unnamed protein product 58.5 2e-06
gi|492501772|ref|WP_005867312.1| hypothetical protein 41.2 0.82
gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 40.4 1.1
gi|575094301|emb|CDL65691.1| unnamed protein product 39.3 4.0
gi|565841291|ref|WP_023924572.1| hypothetical protein 38.9 4.6
>gi|496050831|ref|WP_008775338.1| predicted protein [Bacteroides sp. 2_2_4]
gi|229448895|gb|EEO54686.1| hypothetical protein BSCG_01611 [Bacteroides sp. 2_2_4]
Length=381
Score = 359 bits (921), Expect = 4e-118, Method: Compositional matrix adjust.
Identities = 196/330 (59%), Positives = 254/330 (77%), Gaps = 9/330 (3%)
Query 34 IAQQNNAFNEKMLQKQMDYNTQMYQQQLGDQWQFYNDAKQNSWDMFNAANDYNSASAQRE 93
IAQ NN FNE+MLQKQMDYNT Y QQ+ DQW FYNDAKQN+WDMFNA N+YNSASAQRE
Sbjct 41 IAQMNNEFNERMLQKQMDYNTLAYDQQVSDQWSFYNDAKQNAWDMFNATNEYNSASAQRE 100
Query 94 RLEAAGLNPYLMMSGGNagtataqsspqasspsaqgVTPPTATPYSADYSGITQGLGMVL 153
R EAAGLNPY+MM+ G+AGTA A S+ A++P+ QG+TPPTA+PYSADYSGI QGLG +
Sbjct 101 RYEAAGLNPYVMMNTGSAGTAAATSATSATAPTKQGITPPTASPYSADYSGIMQGLGQAI 160
Query 154 DKIATQPDRDVKSAEADNLRIEGKYKAAKTIAEIVQMRTNAKTQEGRLALDKLIYSIDKD 213
D++++ PD+ AE NL+IEGKYKAA+ IA I ++ + +++ ++AL+KL+YSI KD
Sbjct 161 DQLSSIPDKAKTIAETGNLKIEGKYKAAEAIARIANIKADTHSKKEQVALNKLMYSIQKD 220
Query 214 LKSSQMDVNRESIANMQAERKLTNVQTLLVDKQLSWMDAQSKMDLAQKAADIQLKYAQGA 273
L SS M VN ++IANM+AE K N+QTL+ DKQLS+MDA KM+LA+KAA+IQLK AQGA
Sbjct 221 LASSTMAVNSQNIANMRAEEKFKNIQTLIADKQLSFMDATQKMELAEKAANIQLKLAQGA 280
Query 274 LTRKQVDHEIAKIAETEVRTSLDIQEQTTNVLKQ--------QGMRQENSFNEATFDNRV 325
LTR Q HEI KI+ETE RT+L I EQT+ ++Q Q RQ+N F+ T++ RV
Sbjct 281 LTRNQAAHEIKKISETEARTTL-INEQTSLTIEQNTGQQLQNQAQRQQNRFDADTYNVRV 339
Query 326 KSVKESLWNLMHEADSYGLSKTIGRVIRPL 355
K+++ESL+N++ E D G KT+G+ IR +
Sbjct 340 KTLEESLFNIVFETDKLGAVKTVGKGIRAV 369
>gi|490418711|ref|WP_004291034.1| hypothetical protein [Bacteroides eggerthii]
gi|217986638|gb|EEC52972.1| hypothetical protein BACEGG_02723 [Bacteroides eggerthii DSM
20697]
Length=368
Score = 288 bits (737), Expect = 1e-90, Method: Compositional matrix adjust.
Identities = 165/341 (48%), Positives = 223/341 (65%), Gaps = 30/341 (9%)
Query 28 NKGNASIAQQNNAFNEKMLQKQMDYNTQMYQQQLGDQWQFYNDAKQNSW----------- 76
N+ N IAQ NNAFNEKM KQ+ YN +MYQ QLGDQW+FY+D K N+W
Sbjct 31 NQANKEIAQMNNAFNEKMFDKQIAYNKEMYQTQLGDQWKFYDDQKANAWKLYEDNKAYQT 90
Query 77 DMFNAANDYNSASAQRERLEAAGLNPYLMMSGGN-----agtataqsspqasspsaqgVT 131
+M+N N+YN SAQR RLEAAGLNPY+MM+GG+ + + T S+P A SPSAQGV
Sbjct 91 EMWNKQNEYNDPSAQRARLEAAGLNPYMMMNGGSAGVAGSVSGTQGSAPSAGSPSAQGVQ 150
Query 132 PPTATPYSADYSGITQGLGMVLDKIATQPDRDVKSAEADNLRIEGKYKAAKTIAEIVQMR 191
PPTATPYSADYSG+ QGLG +D I T R++++A+ADNLRIEGKY A+K IAE+ +
Sbjct 151 PPTATPYSADYSGVMQGLGHAIDTIMTGSQRNIQNAQADNLRIEGKYIASKAIAELYKTY 210
Query 192 TNAKTQEGRLALDKLIYSIDKDLKSSQMDVNRESIANMQAERKLTNVQTLLVDKQLSWMD 251
AK + R+A+ +++ SI KDL +SQ+ VN E++ +QA+ K+ + LL ++QL ++
Sbjct 211 NEAKNDDERVAIQRVLSSIQKDLSASQVAVNNENVRQIQAQTKIAVTENLLREQQLKFLP 270
Query 252 AQSKMDLAQKAADIQLKYAQGALTRKQVDHEIAKIAETEVRTSLDIQEQTTNVLKQQGMR 311
+ + LA AADI LKYAQ LT KQ HEI K+AET VR + G
Sbjct 271 YEQRTQLALGAADIALKYAQKNLTEKQARHEIEKLAETIVRAN--------------GQA 316
Query 312 QENSFNEATFDNRVKSVKESLWNLMHEADSYGLSKTIGRVI 352
+N ++ T+ +RVK VKESL+N +++ D G+ KT+ R
Sbjct 317 MQNQYDAETYRDRVKLVKESLFNAIYDTDKVGIFKTMSRAF 357
>gi|547226428|ref|WP_021963491.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103380|emb|CCY83991.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=416
Score = 114 bits (285), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 113/368 (31%), Positives = 174/368 (47%), Gaps = 56/368 (15%)
Query 28 NKGNASIAQQNNAFNEKMLQKQMDYNTQMYQQQLG------DQWQFYNDAKQNSWDMFNA 81
NK N IAQ NN +NE+M KQ++YN M+ QQ+ +Q +N QN + A
Sbjct 30 NKTNLQIAQMNNEYNERMFNKQLEYNQDMFNQQVEYDQKKMEQQNNFNARMQN--EAIGA 87
Query 82 ANDYNSASAQRERLEAAGLNPYLMMSGGNagtataqsspqasspsaqg--VTPPTAT--- 136
YNSA AQR RLEAAGLNPYLMMSGGNAG +A S S S V PPTA+
Sbjct 88 QQVYNSAKAQRARLEAAGLNPYLMMSGGNAGAVSAVSGSSGSGGSPSPMGVNPPTASSAV 147
Query 137 --PYSADYSGITQGLGMVLDKIATQPDRDVKS----AEADNLRIEGKYKAAKTIAEIVQM 190
+ D+SG+T + +LD A + RD ++ +A +IE KYKA K + +I
Sbjct 148 MQAFRPDFSGVTGIIQTLLDIQAQKGVRDAQAFSLGEQASGFKIENKYKAEKLLWDIYNS 207
Query 191 RTNAKTQEGRLALDKLIYSIDKDLKSSQMDVNRESIANMQAERKLTNVQT-------LLV 243
+ + + + +L+ + ++ + + SS + + N Q +L QT LL
Sbjct 208 KADYNLKNSQESLNNMSFARLQAMFSSDVSKAQREAENAQFTGELIRAQTACQQLQGLLG 267
Query 244 DKQLSWMDAQSKMDLAQKAADIQLKYAQGALTRKQVDHEIAKIAETEVRTSLDIQEQTTN 303
K+L + D + +LA +A A G + Q A + +L++ EQ
Sbjct 268 AKELKYYDQKVLQELAIMSAQQYSLVAAGKASEAQ--------ARQAIENALNLVEQ--- 316
Query 304 VLKQQGMRQENSFNEATFDNRVKSVKE----SLWN------------LMHEADSYGLSKT 347
++G++ +N + T + +K+ + S WN + ++ S G +K
Sbjct 317 ---REGIKVDNYVKQKTANALIKTARNNCNTSYWNSKTAHNQSLRPSVFEDSFSQGFNKF 373
Query 348 IGRVIRPL 355
I I PL
Sbjct 374 INTYIAPL 381
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 84.7 bits (208), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 85/319 (27%), Positives = 147/319 (46%), Gaps = 41/319 (13%)
Query 28 NKGNASIAQQNNAFNEKMLQKQMDYNTQMYQQQLGDQWQFYNDAKQNSWDMFNAANDYNS 87
N+ N IAQ +N +N + L++Q++ WDM+NA N+YNS
Sbjct 44 NQANIQIAQMSNEYNREQLERQIE----------------------QEWDMWNAENEYNS 81
Query 88 ASAQRERLEAAGLNPYLMMSGGNagtataqsspqasspsaqgVTPPTATPYSADYSGITQ 147
AS+QR+RLE AGLNPY+MM GG+AG+A++ +SP A + T P AD SG++
Sbjct 82 ASSQRKRLEEAGLNPYMMMDGGSAGSASSMTSPAAQPAVVPQMQGATMQP--ADMSGLSG 139
Query 148 GLGMVLDKIAT-QPDRDVKSAEADN--LRIEGKYKAAKTIAEIVQMRTNAKTQEGRLALD 204
G+ + IAT + D++ + N IE +YKA K +A++ + RT + +
Sbjct 140 LRGIASEFIATLKAQEDIRGQQLINEGQEIENQYKADKLLADLEKTRTESGFVRSQTKGQ 199
Query 205 KLIYSIDKDLKSSQMDVNRESIANMQAERKLTNVQTLLVDKQLSWMDAQSKMDLAQKAAD 264
++ ++ SS++ + Q + L + + Q K + ++
Sbjct 200 DIMNRFRPEMLSSEIRQRKTDTMFTQLRAHGQMLANLSAYQWYKVLPQQIKQTINEQMVR 259
Query 265 IQLKYAQGALTRKQVDHEIAKIAETEVRTSLDIQEQTTNVLKQQGMRQENSFNEATFDNR 324
I QG LT+ Q++ EI K T +K +Q+ F ++ +R
Sbjct 260 INNMKLQGNLTQAQINTEINKA--------------VTEFMKGAREQQQFDFESDSYKDR 305
Query 325 VKSVKESLWNLMHEADSYG 343
+ +K L + ++ + G
Sbjct 306 LDQIKADLRHAIYNSGPEG 324
>gi|575094358|emb|CDL65740.1| unnamed protein product [uncultured bacterium]
Length=328
Score = 74.3 bits (181), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 68/260 (26%), Positives = 124/260 (48%), Gaps = 29/260 (11%)
Query 28 NKGNASIAQQNNAFNEKMLQKQMDYNTQMYQQQLGDQWQFYNDAKQNSWDMFNAANDYNS 87
N N IAQ NN ++E+M++KQM YNT+M+++ DYNS
Sbjct 27 NSTNMQIAQMNNEWSERMMEKQMAYNTEMWEK----------------------VADYNS 64
Query 88 ASAQRERLEAAGLNPYLMMSGGNagtataqsspqasspsaqgVTPPTATPYSADYSGITQ 147
+ ++ AG+NPY+ +SG G+ +A S+ S PS V A P D+S ++
Sbjct 65 LPNKMQQARDAGVNPYMALSGNAFGSISAPSANSVSLPSPSQV---QAQPAQYDFSSVSN 121
Query 148 GL--GMVLDKIA--TQPDRDVKSAEADNLRIEGKYKAAKTIAEIVQMRTNAKTQEGRLAL 203
+ GM L + A + + A D LRIE KY A K ++EI + N K + +
Sbjct 122 SIIAGMDLFQKAQLMKSQQSNIDASTDQLRIENKYHAMKLVSEIAEKMANTKDSQAKAVY 181
Query 204 DKLIYSIDKDLKSSQMDVNRESIANMQAERKLTNVQTLLVDKQLSWMDAQSKMDLAQKAA 263
++I + + +++ ++++NM+ + ++ + +QL + Q + L A+
Sbjct 182 QQIINEYAEQGIKTDLEIKNQTLSNMKETFRGLVLENAMTSEQLRFFPEQVRAQLGLTAS 241
Query 264 DIQLKYAQGALTRKQVDHEI 283
I L + L+++++ I
Sbjct 242 QILLNQSNSKLSQQKMVESI 261
>gi|575094319|emb|CDL65706.1| unnamed protein product [uncultured bacterium]
Length=396
Score = 58.5 bits (140), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 83/336 (25%), Positives = 125/336 (37%), Gaps = 89/336 (26%)
Query 28 NKGNASIAQQNNAFNEKMLQKQMDYNTQMYQQQLGDQWQFYNDAKQNSWDMFNAANDYNS 87
N+ N IA QNN FNE+M +N N+YN
Sbjct 60 NQANREIADQNNKFNERM---------------------------------WNLQNEYNR 86
Query 88 ASAQRERLEAAGLNPYLMMSGGNagtataqsspqasspsaqgVTPPTATPYSADYSGITQ 147
QR RLEAAGLNPYLMM GG+ A S + S + P + Y Q
Sbjct 87 PDMQRARLEAAGLNPYLMMDGGS---AGIAESAPTADTSGTQIAPDIGNTIAGGY----Q 139
Query 148 GLGMVLDKIATQPDRDVKSAEADNLRIEGKYKAAKTIAEIVQMRTNAKTQEGRLALDKLI 207
+G + A+Q + D+L+ K AKT+AE + E R
Sbjct 140 AMGNSISSAASQI---AQMTFQDDLQ---KANVAKTVAEAKNAHLQNQFDELRNEFAVAN 193
Query 208 YSIDKDLKSSQMDVN-------RESIANMQAERK------------------LTNVQTLL 242
+ ++ LK Q D++ R+S+ + K LT+VQ +
Sbjct 194 FLVNLRLKQKQGDISDYEANYLRDSMQDRLDSVKFQNTLSGSQSSYYSQMAGLTDVQRQI 253
Query 243 VDKQLSWMDAQSKMDLAQKAADIQLKYAQGALTRKQVDHEIAKIAETEVRTSLDIQEQTT 302
L W+ + + LA +I+ ++ L Q + A +
Sbjct 254 EQTNLDWLPQEKQAGLAATLQNIRTMVSEMGLNYAQAKNAFAMA--------------SL 299
Query 303 NVLKQQGMRQENSFNEATFDNRVKSVKESL----WN 334
N ++G+R +N E+TFD VK K ++ WN
Sbjct 300 NYANEEGLRIDNRLKESTFDLSVKLAKNTVNSEYWN 335
>gi|492501772|ref|WP_005867312.1| hypothetical protein [Parabacteroides distasonis]
gi|409230405|gb|EKN23269.1| hypothetical protein HMPREF1059_03254 [Parabacteroides distasonis
CL09T03C24]
Length=288
Score = 41.2 bits (95), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 27/70 (39%), Positives = 37/70 (53%), Gaps = 0/70 (0%)
Query 40 AFNEKMLQKQMDYNTQMYQQQLGDQWQFYNDAKQNSWDMFNAANDYNSASAQRERLEAAG 99
A N K +Q N ++ + Q Q Q A Q S +M+N N+YNS + Q R+ AAG
Sbjct 22 AMNNKAVQDTNKANMEIAKYQAQWQQQENEKAYQRSLNMWNLQNEYNSPTQQMARIRAAG 81
Query 100 LNPYLMMSGG 109
LNP L+ G
Sbjct 82 LNPNLVYGNG 91
>gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649557306|gb|KDS63785.1| hypothetical protein M095_3404 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649559158|gb|KDS65545.1| hypothetical protein M096_4689 [Parabacteroides distasonis str.
3999B T(B) 6]
gi|649560567|gb|KDS66875.1| hypothetical protein M095_2448 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561016|gb|KDS67303.1| hypothetical protein M095_2410 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649562727|gb|KDS68911.1| hypothetical protein M096_3341 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=288
Score = 40.4 bits (93), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 27/70 (39%), Positives = 36/70 (51%), Gaps = 0/70 (0%)
Query 40 AFNEKMLQKQMDYNTQMYQQQLGDQWQFYNDAKQNSWDMFNAANDYNSASAQRERLEAAG 99
A N K +Q N ++ + Q Q Q A Q S M+N N+YNS + Q R+ AAG
Sbjct 22 AMNNKAVQDTNKANMEIAKYQAQWQQQENEKAYQRSLKMWNLQNEYNSPTQQMARIRAAG 81
Query 100 LNPYLMMSGG 109
LNP L+ G
Sbjct 82 LNPNLVYGNG 91
>gi|575094301|emb|CDL65691.1| unnamed protein product [uncultured bacterium]
Length=437
Score = 39.3 bits (90), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 16/29 (55%), Positives = 22/29 (76%), Gaps = 0/29 (0%)
Query 78 MFNAANDYNSASAQRERLEAAGLNPYLMM 106
M+ NDYN+ AQ++RLE AG+NPY+ M
Sbjct 69 MWKDTNDYNTPIAQKQRLEQAGMNPYVNM 97
>gi|565841291|ref|WP_023924572.1| hypothetical protein [Prevotella nigrescens]
gi|564729909|gb|ETD29853.1| hypothetical protein HMPREF1173_00035 [Prevotella nigrescens
CC14M]
Length=396
Score = 38.9 bits (89), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 49/209 (23%), Positives = 88/209 (42%), Gaps = 32/209 (15%)
Query 26 AGNKGNASIAQQNNAFNEKMLQKQMDYNTQMYQQQLGDQWQFYNDAKQNSWDMFNAANDY 85
+ N N IA++ NA N +M+Q Q ++N +M +Q N+Y
Sbjct 29 SANSTNLRIARETNAANFQMMQYQNEFNQKMLDKQ----------------------NEY 66
Query 86 NSASAQRERLEAAGLNPYLMMSGGNagtataqsspqasspsaqgVTPPTATPYSADYSGI 145
QR+R E AG+NPY +S ++GT P+ P A +
Sbjct 67 ALPINQRKRFEDAGINPYFALSQISSGTPQGALQSAQGHPAVAAQVQPVTAFGDALRDSV 126
Query 146 TQGL---GMVLDKIATQPDRDVKSAEADNLRIEGKYKAAKTIAEIVQMRTNAKTQEGRLA 202
+ G+ G ++ TQ +A+ +E ++KAA ++ I + K+ +
Sbjct 127 SHGVNTYGQLMQAKYTQQ-------QAEGQSLENRFKAATLLSRIDGEKAKNKSLTYNMM 179
Query 203 LDKLIYSIDKDLKSSQMDVNRESIANMQA 231
+D L + K + ++M + S+A M+A
Sbjct 180 MDGLRADLMKYVNGNEMKKSDLSVAQMEA 208
Lambda K H a alpha
0.311 0.125 0.341 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2134211136096