bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-39_CDS_annotation_glimmer3.pl_2_1
Length=225
Score E
Sequences producing significant alignments: (Bits) Value
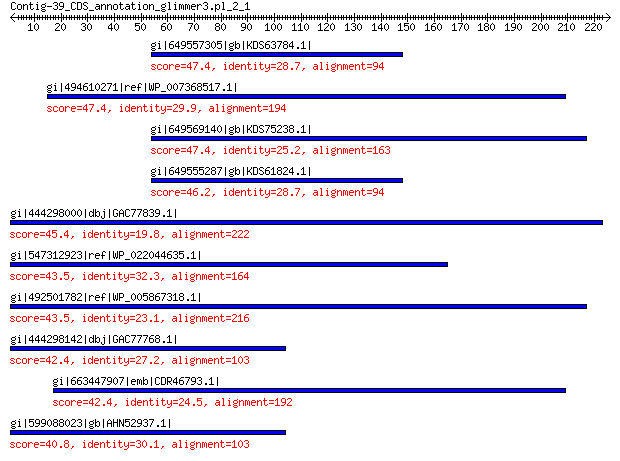
gi|649557305|gb|KDS63784.1| capsid family protein 47.4 0.002
gi|494610271|ref|WP_007368517.1| capsid protein 47.4 0.004
gi|649569140|gb|KDS75238.1| capsid family protein 47.4 0.004
gi|649555287|gb|KDS61824.1| capsid family protein 46.2 0.009
gi|444298000|dbj|GAC77839.1| major capsid protein 45.4 0.015
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 43.5 0.058
gi|492501782|ref|WP_005867318.1| hypothetical protein 43.5 0.077
gi|444298142|dbj|GAC77768.1| major capsid protein 42.4 0.16
gi|663447907|emb|CDR46793.1| CYFA0S26e00166g1_1 42.4 0.17
gi|599088023|gb|AHN52937.1| major capsid protein 40.8 0.37
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 47.4 bits (111), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 50/95 (53%), Gaps = 2/95 (2%)
Query 54 TYYFKEPGYIFDMMTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYGW 112
T YF+E GYI +M+IRP + G+ D+ ++ D++ P + +G Q++ L Y
Sbjct 92 TRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEL-YLN 150
Query 113 KADTVSSLSVAKEPCYNEFRSSYDEVLGSLQATLT 147
++D + + P Y E++ S +EV G + +
Sbjct 151 ESDAANEGTFGYTPRYAEYKYSQNEVHGDFRGNMA 185
>gi|494610271|ref|WP_007368517.1| capsid protein [Prevotella multiformis]
gi|324988543|gb|EGC20506.1| putative capsid protein (F protein) [Prevotella multiformis DSM
16608]
Length=531
Score = 47.4 bits (111), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 58/226 (26%), Positives = 96/226 (42%), Gaps = 40/226 (18%)
Query 15 NSQVVLNQAGQSGFEGG--ESAALGQMGGSISFNTVLGREQTYYFKEPGYIFDMMTIRPV 72
N V+ QS F+ G ES LG +GG ++ + KE G I + ++ P
Sbjct 300 NPVVISEVVNQSEFDRGADESPCLGDLGGK-GVGSLNSSSIDFDVKEHGIIMCIYSVVPQ 358
Query 73 YFWTG--IRPDYLEYRGPDYFNPIYNDIGYQ------------DVPL-------WRLGYG 111
+ G P + R D+F P + D+GYQ D P+ RL G
Sbjct 359 TEYNGTYFDPFNRKLRREDFFQPEFADLGYQPVVTSDLISTYLDNPVPDGPEKQKRLAAG 418
Query 112 WKADTVSSLS--VAKEPCYNEFRSSYDEVLGSLQATLTPKASTPLQSYWVQQR-DFYLIG 168
+ ++ + + + + YNE+++S D V G ++ L+ SYW R DF G
Sbjct 419 YPLSSIEANNRLLGWQVRYNEYKTSRDLVFGEFESGLS-------LSYWCSPRYDFGFDG 471
Query 169 LSSNPNEV----SPSMLFTNLNTVNNPF--ASDMEDNFFVNMSYKV 208
+ + V SP+ + N + +N F ++ D+F VN + V
Sbjct 472 KAGDKKLVNSPWSPAHFYVNPSILNTIFLVSAVKADHFLVNSFFDV 517
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 47.4 bits (111), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 41/166 (25%), Positives = 74/166 (45%), Gaps = 21/166 (13%)
Query 54 TYYFKEPGYIFDMMTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYGW 112
T YF+E GYI +M+IRP + G+ D+ ++ D++ P + +G Q++ L Y
Sbjct 237 TRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEL-YLN 295
Query 113 KADTVSSLSVAKEPCYNEFRSSYDEVLGSLQATLTPKASTPLQSYWVQQRDFYLIGLSSN 172
++D + + P Y E++ S +EV G + + ++W R F
Sbjct 296 ESDAANEGTFGYTPRYAEYKYSQNEVHGDFRGNM---------AFWHLNRIF-----KEK 341
Query 173 PNEVSPSMLFTNLNTVNNPFAS--DMEDNFFVNMSYKVVVKNLINK 216
PN + F N N FA+ +D ++V + + L+ K
Sbjct 342 PNL---NTTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPK 384
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 46.2 bits (108), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 50/95 (53%), Gaps = 2/95 (2%)
Query 54 TYYFKEPGYIFDMMTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYGW 112
T YF+E GYI +M+IRP + G+ D+ ++ D++ P + +G Q++ L Y
Sbjct 388 TRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEL-YLN 446
Query 113 KADTVSSLSVAKEPCYNEFRSSYDEVLGSLQATLT 147
++D + + P Y E++ S +EV G + +
Sbjct 447 ESDAANEGTFGYTPRYAEYKYSQNEVHGDFRGNMA 481
>gi|444298000|dbj|GAC77839.1| major capsid protein [uncultured marine virus]
Length=480
Score = 45.4 bits (106), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 44/227 (19%), Positives = 89/227 (39%), Gaps = 29/227 (13%)
Query 1 VDRPKLLFSSSVMVNSQVVLNQAGQSGFEGGESAALGQMGGSISFNTVLGREQTY--YFK 58
+ RP+ + + +N VL + + G + + +G R Y Y +
Sbjct 277 LQRPEYMGGGTTQINFSEVLQTSPE--IPGEDQVSQFGVGDMYGHGIAAMRSNKYRRYIE 334
Query 59 EPGYIFDMMTIRPVYFWT-GIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYGWKADTV 117
E GYI M+++RP +T GI +L DY+ IG Q++ + + +
Sbjct 335 EHGYIISMLSVRPKTMYTNGIHRSWLRLTKEDYYQKELEHIGQQEIMNNEI---YADEGA 391
Query 118 SSLSVAKEPCYNEFRSSYDEVLGSLQATLTPKASTPLQSYWVQQRDFYLIGLSSNPNEVS 177
+ + Y+E+R + V + L +YW R+F E
Sbjct 392 GTETFGYNDRYSEYRETPSHVSAEFRGIL---------NYWHMAREF----------EAP 432
Query 178 PSM--LFTNLNTVNNPFASDMEDNFFVNMSYKVVVKNLINKSFATRL 222
P + F + + +D ++ + +K+V + L++++ A R+
Sbjct 433 PVLNQSFVDCDATKRIHNEQTQDALWIMIQHKMVARRLLSRNAAPRI 479
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 43.5 bits (101), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 53/192 (28%), Positives = 74/192 (39%), Gaps = 40/192 (21%)
Query 1 VDRPKLLFSSSVMVNSQVVLNQAGQSGFEGGESAALGQMGGSIS----FNTVLGREQTYY 56
V++P L +N V +A +G GE A LGQ+ + F+ G + YY
Sbjct 94 VNKPDFLGVWQASINPSNV--RAMANGSASGEDANLGQLAACVDRYCDFSGHSGID--YY 149
Query 57 FKEPG--YIFDMMTIRPVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRL-----G 109
KEPG + M+ P Y G+ PD D FNP N IG+Q VP R G
Sbjct 150 AKEPGTFMLITMLVPEPAYS-QGLHPDLASISFGDDFNPELNGIGFQLVPRHRFSMMPRG 208
Query 110 YG----------WKADTVSS-------LSVAKEPCYNEFRSSYDEVLGSLQATLTPKAST 152
+ W T + +SV +E ++ R+ Y + G A
Sbjct 209 FNFTGLDQEASPWFGHTGTGVLVDPNMVSVGEEVAWSWLRTDYSRLHGDF-------AQN 261
Query 153 PLQSYWVQQRDF 164
YWV R F
Sbjct 262 GNYQYWVLTRRF 273
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 43.5 bits (101), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 50/219 (23%), Positives = 92/219 (42%), Gaps = 27/219 (12%)
Query 1 VDRPKLLFSSSVMVNSQVVLNQAGQSGFEGGESAALGQMGGSISFNTVLGREQTYYFKEP 60
+ RP+ L ++ VL Q+ S G IS G ++ YF+E
Sbjct 338 LQRPQFLGGGRTPISVSEVL----QTSATDSTSPQANMAGHGISAGVNHGFKR--YFEEH 391
Query 61 GYIFDMMTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYGWKADTVSS 119
GYI +M+IRP + G+ D+ ++ D++ P + +G Q++ + Y + ++
Sbjct 392 GYIIGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEV-YLQQTPASNN 450
Query 120 LSVAKEPCYNEFRSSYDEVLGSLQATLTPKASTPLQSYWVQQRDFYLIGLSSNPNEVSPS 179
+ P Y E++ S +EV G + + ++W R F S +PN +
Sbjct 451 GTFGYTPRYAEYKYSMNEVHGDFRGNM---------AFWHLNRIF-----SESPNL---N 493
Query 180 MLFTNLNTVNNPFAS--DMEDNFFVNMSYKVVVKNLINK 216
F N N FA+ +D +++ + V L+ K
Sbjct 494 TTFVECNPSNRVFATAETSDDKYWIQLYQDVKALRLMPK 532
>gi|444298142|dbj|GAC77768.1| major capsid protein [uncultured marine virus]
Length=299
Score = 42.4 bits (98), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 28/104 (27%), Positives = 47/104 (45%), Gaps = 5/104 (5%)
Query 1 VDRPKLLFSSSVMVNSQVVLNQAGQSGFEGGESAALGQMGGSISFNTVLGREQTYYFKEP 60
+ RP+++ + +N VLN G SG + LG+MGG V Y+ +E
Sbjct 160 LQRPEMISTGKSNINFSEVLNTTGPSGV---DDHPLGEMGGH-GIAGVKSNRARYFCEEH 215
Query 61 GYIFDMMTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDV 103
G+I +M++RP + T + DY+ IG ++V
Sbjct 216 GHIISLMSVRPKTIYMTTQHKQFDRESKEDYWQKELQAIGMEEV 259
>gi|663447907|emb|CDR46793.1| CYFA0S26e00166g1_1 [Cyberlindnera fabianii]
Length=388
Score = 42.4 bits (98), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 47/213 (22%), Positives = 88/213 (41%), Gaps = 39/213 (18%)
Query 17 QVVLNQAGQSGFEGGESAALGQMGGSISFNTVLGREQTYYFKEPGYI---FDMMTIRPVY 73
Q+++ Q G + F GE+AA+G ++ T Y KEPGY+ D +
Sbjct 80 QILMRQRG-TKFYPGENAAIG-------------KDHTIYAKEPGYVRFYLDPFHPNRKF 125
Query 74 FWTGIRPDYLEYRGPD-YFNPIYNDIGY-----------QDVPLWRLGYGWKADTVSSLS 121
+RPD R P +F P +GY ++ L R + + + + SL
Sbjct 126 IGVALRPD---LRLPTAHFEPSIRRLGYVPITDAKKAQFEENNLSRKAHLMRPEIIKSLK 182
Query 122 VAKEPCYNEFRSSYDEVLGSLQATLTPKASTPLQSYWVQQRDFYLIGLSSNPNEVSPSML 181
+E E S+Y + S+ A LT S + + R+ GL N + + + +
Sbjct 183 -QREAKRQELLSTYSSQITSIVADLTESDSQLAAARLLSIRNHIKAGLPLNQAQATTTSV 241
Query 182 FTNLNTVNNPFA------SDMEDNFFVNMSYKV 208
+ + +++ +D N ++++S K+
Sbjct 242 YLHDLKLDSKKGLMTTEDADASKNAYISLSNKI 274
>gi|599088023|gb|AHN52937.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=213
Score = 40.8 bits (94), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 31/104 (30%), Positives = 48/104 (46%), Gaps = 9/104 (9%)
Query 1 VDRPKLLFSSSVMVNSQVVLNQAGQSGFEGGESAALGQMGGSISFNTVLGREQTYYFKEP 60
+ RP+ + S MVN V N AGQSG G+ A+G + GS + TY E
Sbjct 112 IQRPEYIGGGSSMVNVTPVANTAGQSGDYVGQLGAMGTVSGS--------HDWTYSAVEH 163
Query 61 GYIFDMMTIR-PVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDV 103
G I + +R + + G+ + + D++ P+ IG Q V
Sbjct 164 GVIIGLANVRGDITYSQGLERYWSKSTRYDFYYPVLAQIGEQAV 207
Lambda K H a alpha
0.318 0.135 0.406 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 880107843099