bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-38_CDS_annotation_glimmer3.pl_2_1
Length=229
Score E
Sequences producing significant alignments: (Bits) Value
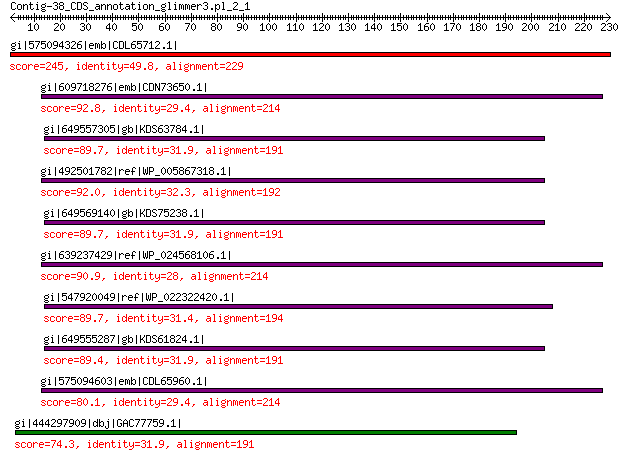
gi|575094326|emb|CDL65712.1| unnamed protein product 245 2e-72
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 92.8 3e-18
gi|649557305|gb|KDS63784.1| capsid family protein 89.7 3e-18
gi|492501782|ref|WP_005867318.1| hypothetical protein 92.0 5e-18
gi|649569140|gb|KDS75238.1| capsid family protein 89.7 1e-17
gi|639237429|ref|WP_024568106.1| hypothetical protein 90.9 1e-17
gi|547920049|ref|WP_022322420.1| capsid protein VP1 89.7 4e-17
gi|649555287|gb|KDS61824.1| capsid family protein 89.4 4e-17
gi|575094603|emb|CDL65960.1| unnamed protein product 80.1 5e-14
gi|444297909|dbj|GAC77759.1| major capsid protein 74.3 9e-13
>gi|575094326|emb|CDL65712.1| unnamed protein product [uncultured bacterium]
Length=758
Score = 245 bits (626), Expect = 2e-72, Method: Compositional matrix adjust.
Identities = 114/229 (50%), Positives = 165/229 (72%), Gaps = 5/229 (2%)
Query 1 MQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGV 60
++GR+D+++R+D L MPE++GGI+R++ + + QTV+ T+ G Y +LGS+SG+A
Sbjct 535 IEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVE---TTGSGSYVGSLGSQSGLATC 591
Query 61 YGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPIT 120
+G+T +I VFCDEES ++G++ V P+P+Y +L K Y LD + PEFD IG+QPI
Sbjct 592 FGNTDGSISVFCDEESIVMGIMYVMPMPVYDSLLPKWLTYRERLDSFNPEFDHIGYQPIY 651
Query 121 YKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQ 180
KE+ P+ V D ++ N FGYQRPWYEYVAK D AHGLF ++++NF+M R F +P+
Sbjct 652 AKELGPMQC-VQDDID-PNTVFGYQRPWYEYVAKPDRAHGLFLSSLRNFIMFRSFDNVPE 709
Query 181 LGQQFLLVDPDTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 229
LGQ F ++ P +VN VFSVTE +DKI G + F+ TA+LPISRV +PRL+
Sbjct 710 LGQSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPISRVVVPRLE 758
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 92.8 bits (229), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 63/216 (29%), Positives = 105/216 (49%), Gaps = 14/216 (6%)
Query 13 ELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFC 72
L PEF+GG + + V Q ST+ QG A G GI G + F
Sbjct 330 RLQRPEFLGGNKSPIMISEVLQQSATDSTTPQGNMA---GHGIGIGKDGGFSR-----FF 381
Query 73 DEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVA 132
+E Y+IGL++V P Y+Q + + F + D++ P+F+ IG QP+ KE+ N+
Sbjct 382 EEHGYVIGLMSVIPKTSYSQGIPRHFSKSDKFDYFWPQFEHIGEQPVYNKEIFAKNIDAF 441
Query 133 DTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVF--SGLPQLGQQFLLVDP 190
D+ FGY + EY + HG F+ ++ + + R+F P L Q F+ D
Sbjct 442 DS----EAVFGYLPRYSEYKFSPSTVHGDFKDDLYFWHLGRIFDTDKPPVLNQSFIECDK 497
Query 191 DTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIP 226
+ ++++F+V + TDK + ++ TA+ +S P
Sbjct 498 NALSRIFAVEDDTDKFYCHLYQKITAKRKMSYFGDP 533
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 61/191 (32%), Positives = 92/191 (48%), Gaps = 15/191 (8%)
Query 14 LLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCD 73
L P+F+GG +S+ V QT STS Q A G+ ++ + +
Sbjct 45 LQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAGH--------GISAGVNHGFTRYFE 96
Query 74 EESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVAD 133
E YI+G++++ P Y Q + KDF +D Y PEF +G Q I +E L L +D
Sbjct 97 EHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEE---LYLNESD 153
Query 134 TVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTV 193
N+ TFGY + EY + HG FR NM + ++R+F P L F+ +P
Sbjct 154 AANEG--TFGYTPRYAEYKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPS-- 209
Query 194 NQVFSVTEYTD 204
N+VF+ E +D
Sbjct 210 NRVFATAETSD 220
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 92.0 bits (227), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 62/192 (32%), Positives = 92/192 (48%), Gaps = 15/192 (8%)
Query 13 ELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFC 72
L P+F+GG +S+ V QT STS Q A G+ ++ + +
Sbjct 337 RLQRPQFLGGGRTPISVSEVLQTSATDSTSPQANMAGH--------GISAGVNHGFKRYF 388
Query 73 DEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVA 132
+E YIIG++++ P Y Q + KDF +D Y PEF +G Q I +EV +
Sbjct 389 EEHGYIIGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEVY-----LQ 443
Query 133 DTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDT 192
T N TFGY + EY + HG FR NM + ++R+FS P L F+ +P
Sbjct 444 QTPASNNGTFGYTPRYAEYKYSMNEVHGDFRGNMAFWHLNRIFSESPNLNTTFVECNPS- 502
Query 193 VNQVFSVTEYTD 204
N+VF+ E +D
Sbjct 503 -NRVFATAETSD 513
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 89.7 bits (221), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 61/191 (32%), Positives = 92/191 (48%), Gaps = 15/191 (8%)
Query 14 LLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCD 73
L P+F+GG +S+ V QT STS Q A G+ ++ + +
Sbjct 190 LQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAGH--------GISAGVNHGFTRYFE 241
Query 74 EESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVAD 133
E YI+G++++ P Y Q + KDF +D Y PEF +G Q I +E L L +D
Sbjct 242 EHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEE---LYLNESD 298
Query 134 TVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTV 193
N+ TFGY + EY + HG FR NM + ++R+F P L F+ +P
Sbjct 299 AANEG--TFGYTPRYAEYKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPS-- 354
Query 194 NQVFSVTEYTD 204
N+VF+ E +D
Sbjct 355 NRVFATAETSD 365
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 90.9 bits (224), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 60/216 (28%), Positives = 103/216 (48%), Gaps = 14/216 (6%)
Query 13 ELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFC 72
L PEF+GG + + V Q ST+ QG A G+ F
Sbjct 339 RLQRPEFLGGNKTPILISEVLQQSSTDSTTPQGNMAGH--------GISVGKEGGFSKFF 390
Query 73 DEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVA 132
+E Y+IGL++V P Y+Q + + F D++ P+F+ IG QP+ KE+ N+G
Sbjct 391 EEHGYVIGLMSVIPKTSYSQGIPRHFSKFDKFDYFWPQFEHIGEQPVYNKEIFAKNVGDY 450
Query 133 DTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVF--SGLPQLGQQFLLVDP 190
D+ FGY + EY + HG F+ + + + R+F S P+L + F+ V+
Sbjct 451 DS----GGVFGYVPRYSEYKYSPSTIHGDFKDTLYFWHLGRIFDSSAPPKLNRDFIEVNK 506
Query 191 DTVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIP 226
++++F+V + +DK + ++ TA+ +S P
Sbjct 507 SGLSRIFAVEDNSDKFYCHLYQKITAKRKMSYFGDP 542
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 89.7 bits (221), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 61/194 (31%), Positives = 91/194 (47%), Gaps = 15/194 (8%)
Query 14 LLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCD 73
L P+F+GG +S+ V QT TS Q A G+ +N + + +
Sbjct 353 LQRPQFLGGGRMPISVSEVLQTSSTDETSPQANMAGH--------GISAGINNGFKHYFE 404
Query 74 EESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVAD 133
E YIIG++++TP Y Q + +DF +D Y PEF + Q I +E L V++
Sbjct 405 EHGYIIGIMSITPRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQEIKNQE-----LFVSE 459
Query 134 TVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTV 193
N TFGY + EY AHG FR N+ + ++R+F P L F+ P
Sbjct 460 DAAYNNGTFGYTPRYAEYKYHPSEAHGDFRGNLSFWHLNRIFEDKPNLNTTFVECKPS-- 517
Query 194 NQVFSVTEYTDKIF 207
N+VF+ +E D F
Sbjct 518 NRVFATSETEDDKF 531
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 89.4 bits (220), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 61/191 (32%), Positives = 92/191 (48%), Gaps = 15/191 (8%)
Query 14 LLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCD 73
L P+F+GG +S+ V QT STS Q A G+ ++ + +
Sbjct 341 LQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAGH--------GISAGVNHGFTRYFE 392
Query 74 EESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVAD 133
E YI+G++++ P Y Q + KDF +D Y PEF +G Q I +E L L +D
Sbjct 393 EHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEE---LYLNESD 449
Query 134 TVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDTV 193
N+ TFGY + EY + HG FR NM + ++R+F P L F+ +P
Sbjct 450 AANEG--TFGYTPRYAEYKYSQNEVHGDFRGNMAFWHLNRIFKEKPNLNTTFVECNPS-- 505
Query 194 NQVFSVTEYTD 204
N+VF+ E +D
Sbjct 506 NRVFATAETSD 516
>gi|575094603|emb|CDL65960.1| unnamed protein product [uncultured bacterium]
Length=507
Score = 80.1 bits (196), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 63/215 (29%), Positives = 100/215 (47%), Gaps = 20/215 (9%)
Query 13 ELLMPEFIGGISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFC 72
L PE++GG R + V QT + S +G+ G G+ + F
Sbjct 311 RLQNPEYLGGGQRTIQFSEVLQTSEGSS---------PVGTLRG-HGIGALKTKRFLRFF 360
Query 73 DEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGFQPITYKEVCPLNLGVA 132
+E ++ LL V P+ IYTQ L++ FL D++QP+F+ IG Q + KE+ A
Sbjct 361 NEHGILLTLLVVRPISIYTQGLNRMFLRETRFDYWQPQFEHIGQQSVLNKEL------YA 414
Query 133 DTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFSGLPQLGQQFLLVDPDT 192
+ N + TFG+ + EY + HG FRT +++ + R F P L FL P T
Sbjct 415 ASPN-GDDTFGFSNRYNEYRYHPSNVHGEFRTYYEDWHLGRKFESTPTLNSDFLKCHPTT 473
Query 193 VNQVFSVTEYT-DKIFGYVKFNATARLPISRVAIP 226
++F+ T D++ + + AR IS+ P
Sbjct 474 --RIFAETSGNYDQLLVMCQNHIRARRLISKNGDP 506
>gi|444297909|dbj|GAC77759.1| major capsid protein, partial [uncultured marine virus]
Length=257
Score = 74.3 bits (181), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 61/197 (31%), Positives = 89/197 (45%), Gaps = 22/197 (11%)
Query 3 GRWDIDIRFDELLMPEFIGG------ISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSG 56
W + L PE++GG +S LS T E +DQ+ + Q A G
Sbjct 74 AHWGVRSSDARLDRPEYLGGGKQPVLVSEVLS--TAEVAIDQEISIPQANMA---GHGIS 128
Query 57 IAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRIGF 116
+ G SN + +E +I+G+++V P Y Q + + F D+Y PEF +G
Sbjct 129 VGG-----SNRFKKRFEEHGHILGIMSVIPRTAYQQGVDRSFSREDKFDYYFPEFAHLGE 183
Query 117 QPITYKEVCPLNLGVADTVNKANQTFGYQRPWYEYVAKYDSAHGLFRTNMKNFVMSRVFS 176
Q + EV +G DT N + TFGYQ + EY K G FR N+ + M R F+
Sbjct 184 QSVNNYEVY---MG-DDTEN--HDTFGYQSRYAEYKYKNSMVTGDFRDNLDFWHMGRQFA 237
Query 177 GLPQLGQQFLLVDPDTV 193
P LG +F+ P V
Sbjct 238 TRPVLGDEFVQSKPTXV 254
Lambda K H a alpha
0.321 0.138 0.409 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 907704005640