bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-35_CDS_annotation_glimmer3.pl_2_1
Length=174
Score E
Sequences producing significant alignments: (Bits) Value
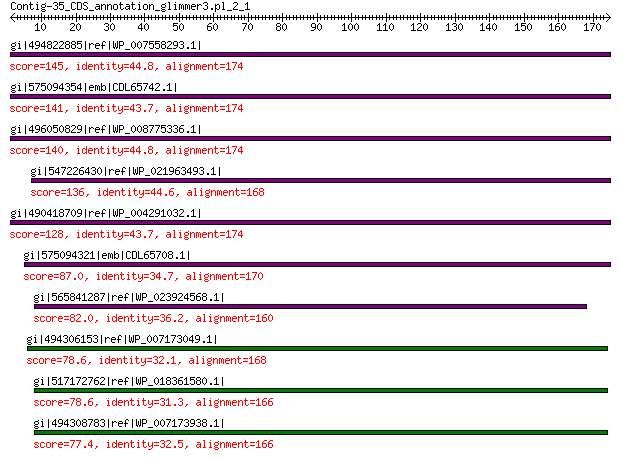
gi|494822885|ref|WP_007558293.1| hypothetical protein 145 9e-38
gi|575094354|emb|CDL65742.1| unnamed protein product 141 6e-36
gi|496050829|ref|WP_008775336.1| hypothetical protein 140 6e-36
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 136 2e-34
gi|490418709|ref|WP_004291032.1| hypothetical protein 128 2e-31
gi|575094321|emb|CDL65708.1| unnamed protein product 87.0 1e-16
gi|565841287|ref|WP_023924568.1| hypothetical protein 82.0 6e-15
gi|494306153|ref|WP_007173049.1| hypothetical protein 78.6 6e-14
gi|517172762|ref|WP_018361580.1| hypothetical protein 78.6 7e-14
gi|494308783|ref|WP_007173938.1| hypothetical protein 77.4 2e-13
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 145 bits (367), Expect = 9e-38, Method: Compositional matrix adjust.
Identities = 78/183 (43%), Positives = 104/183 (57%), Gaps = 9/183 (5%)
Query 1 MTYNTGSKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLV 60
+ +N G +Y I+MC++H +P LDY S +TN+ D PIPEFD IGME VPV++ +
Sbjct 431 INFNVGGQYGIVMCVFHVLPQLDYITSAPHFGTTLTNVLDFPIPEFDKIGMEQVPVIRGL 490
Query 61 NSSLLSTNIGKYS---FFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWLPL 117
N K S +FGY P+YYNWKT +D+ G F LK W+ P DD L +
Sbjct 491 NPVKPKDGDFKVSPNLYFGYAPQYYNWKTTLDKSMGEFRRSLKTWIIPFDDEALLAADSV 550
Query 118 ------TSSQPSLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDG 171
S+ FFKV+P+ LDN+FAVKA+S TDQ L + D VVR L +G
Sbjct 551 DFPDNPNVEADSVKAGFFKVSPSVLDNLFAVKANSDLNTDQFLCSTLFDVNVVRSLDPNG 610
Query 172 IPY 174
+PY
Sbjct 611 LPY 613
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 141 bits (355), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 76/180 (42%), Positives = 102/180 (57%), Gaps = 6/180 (3%)
Query 1 MTYNTGSKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLV 60
+ + + +Y IIMCIYH +P++DY SG D + + PIPE D IGME+VP+V+ +
Sbjct 436 IRFESKGEYGIIMCIYHVLPIVDYVGSGVDHSCTLVDATSFPIPELDQIGMESVPLVRAM 495
Query 61 NSSLLSTNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWLPLT-S 119
N S +F GY PRY +WKT +DR G F L+ W P+ D L L
Sbjct 496 NPVKESDTPSADTFLGYAPRYIDWKTSVDRSVGDFADSLRTWCLPVGDKELTSANSLNFP 555
Query 120 SQP-----SLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIPY 174
S P S+ FFKVNP+ +D +FAV ADST TD+ L + D KVVR L +G+PY
Sbjct 556 SNPNVEPDSIAAGFFKVNPSIVDPLFAVVADSTVKTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 140 bits (353), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 78/179 (44%), Positives = 104/179 (58%), Gaps = 6/179 (3%)
Query 1 MTYNTGSKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLV 60
++++ G +Y +IMCIYH++PLLDY + N D IPEFD +GME+VP+V L+
Sbjct 403 ISFDAGERYGLIMCIYHSLPLLDYTTDLVNPAFTKINSTDFAIPEFDRVGMESVPLVSLM 462
Query 61 NSSLLSTNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWL----- 115
N S N+G S GY PRY ++KT +D GAF T LK WV D+ + L
Sbjct 463 NPLQSSYNVGS-SILGYAPRYISYKTDVDSSVGAFKTTLKSWVMSYDNQSVINQLNYQDD 521
Query 116 PLTSSQPSLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIPY 174
P S + Y FKVNPN +D +FAV A ++ TDQ L + D KVVR L DG+PY
Sbjct 522 PNNSPGTLVNYTNFKVNPNCVDPLFAVAASNSIDTDQFLCSSFFDVKVVRNLDTDGLPY 580
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 136 bits (342), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 75/173 (43%), Positives = 103/173 (60%), Gaps = 6/173 (3%)
Query 7 SKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAV-PVVQLVNSSLL 65
S++ IIMCIYH +PLLD++I+ Q T D IPEFD++GM+ + P + L
Sbjct 402 SEHGIIMCIYHCLPLLDWSINRIARQNFKTTFTDYAIPEFDSVGMQQLYPSEMIFGLEDL 461
Query 66 STNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWLPLTS----SQ 121
++ + GY PRY + KT ID +HG+F L WV+P+ DSY+ + S
Sbjct 462 PSDPSSINM-GYVPRYADLKTSIDEIHGSFIDTLVSWVSPLTDSYISAYRQACKDAGFSD 520
Query 122 PSLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIPY 174
++ Y FFKVNP+ +DNIF VKADST TDQLL+N D K VR +G+PY
Sbjct 521 ITMTYNFFKVNPHIVDNIFGVKADSTINTDQLLINSYFDIKAVRNFDYNGLPY 573
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 128 bits (321), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 76/186 (41%), Positives = 102/186 (55%), Gaps = 13/186 (7%)
Query 1 MTYNTGSKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLV 60
+ +N+ +Y +IMCIYH +PLLDY D L N D IPEFD +GM+++P+VQL+
Sbjct 394 INFNSNGRYGLIMCIYHCLPLLDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPLVQLM 453
Query 61 NSSLLSTNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDD-SYLQQWL---- 115
N L S GY PRY ++KT +D+ G F L WV + S L+Q
Sbjct 454 N-PLRSFANASGLVLGYVPRYIDYKTSVDQSVGGFKRTLNSWVISYGNISVLKQVTLPND 512
Query 116 --PLTSSQP-----SLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLS 168
P+ S+P + + FFKVNP+ LD IFAV+A TDQ L + D K VR L
Sbjct 513 APPIEPSEPVPSVAPMNFTFFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFDIKAVRNLD 572
Query 169 QDGIPY 174
DG+PY
Sbjct 573 TDGLPY 578
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 87.0 bits (214), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 59/186 (32%), Positives = 87/186 (47%), Gaps = 22/186 (12%)
Query 5 TGSKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLV---- 60
T Y +++ IY P+LD+A G D L T+ D IPE D+IGM+ ++
Sbjct 461 TAKTYGVVIGIYRCTPVLDFAHLGIDRTLFKTDASDFVIPEMDSIGMQQTFRCEVAAPAP 520
Query 61 -NSSLLSTNIGKYS------FFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQ 113
N + +G S +GY PRY +KT DR +GAF LK WV I+ +Q
Sbjct 521 YNDEFKAFRVGDGSSPDMSETYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGINFDAIQN 580
Query 114 -----WLPLTSSQPSLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLS 168
W + + P++ F P+ + N+F V + + DQL V C R LS
Sbjct 581 NVWNTWAGINA--PNM----FACRPDIVKNLFLVSSTNNSDDDQLYVGMVNMCYATRNLS 634
Query 169 QDGIPY 174
+ G+PY
Sbjct 635 RYGLPY 640
>gi|565841287|ref|WP_023924568.1| hypothetical protein [Prevotella nigrescens]
gi|564729907|gb|ETD29851.1| hypothetical protein HMPREF1173_00033 [Prevotella nigrescens
CC14M]
Length=656
Score = 82.0 bits (201), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 58/167 (35%), Positives = 87/167 (52%), Gaps = 17/167 (10%)
Query 8 KYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQ-----LVNS 62
++ +IMCIY P +DY D + ED PEF+N+GM+ PV+Q +NS
Sbjct 492 EHGLIMCIYSIAPQVDYDARELDPFNRKFSREDYFQPEFENLGMQ--PVIQSDLCLCINS 549
Query 63 SLLSTNIGKYSFFGYNPRYYNWKTKIDRVHGAFTT--DLKDWVAPIDDSYLQQWLPLTSS 120
+ ++ + GY+ RY +KT D + G F + L W P ++Y ++ L S
Sbjct 550 AKSDSSDQHNNVLGYSARYLEYKTARDIIFGEFMSGGSLSAWATP-KNNYTFEFGKL--S 606
Query 121 QPSLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPL 167
P LL V+P L+ IFAVK + + +TDQ LVN D K +RP+
Sbjct 607 LPDLL-----VDPKVLEPIFAVKYNGSMSTDQFLVNSYFDVKAIRPM 648
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 78.6 bits (192), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 54/173 (31%), Positives = 84/173 (49%), Gaps = 19/173 (11%)
Query 6 GSKYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLVNSSLL 65
++ ++MCIY +P + Y + D + + D PEF+N+GM Q +NSS +
Sbjct 359 AKEHGVLMCIYSLVPQIQYDCTRLDPMVDKLDRFDFFTPEFENLGM------QPLNSSYI 412
Query 66 S---TNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTD--LKDWVAPIDDSYLQQWLPLTSS 120
S T K GY PRY +KT +D HG F + L W S ++W
Sbjct 413 SSFCTPDPKNPVLGYQPRYSEYKTALDINHGQFAQNDALSSWSV----SRFRRWTTF--- 465
Query 121 QPSLLYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIP 173
P L FK++P L+++F V+ + T +TD + CN + V +S DG+P
Sbjct 466 -PQLEIADFKIDPGCLNSVFPVEFNGTESTDCVFGGCNFNIVKVSDMSVDGMP 517
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 78.6 bits (192), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 52/169 (31%), Positives = 80/169 (47%), Gaps = 10/169 (6%)
Query 8 KYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGME---AVPVVQLVNSSL 64
++ I+MCIY +P + Y D + D +PEF+N+GM+ A + N++
Sbjct 405 EHGILMCIYSLVPDVQYDSKRVDPFVQKIERGDFFVPEFENLGMQPLFAKNISYKYNNNT 464
Query 65 LSTNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWLPLTSSQPSL 124
++ I FG+ PRY +KT +D HG F P+ SY S +
Sbjct 465 ANSRIKNLGAFGWQPRYSEYKTALDINHGQFVHQ-----EPL--SYWTVARARGESMSNF 517
Query 125 LYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIP 173
FK+NP LD++FAV + T TDQ+ C + V +S DG+P
Sbjct 518 NISTFKINPKWLDDVFAVNYNGTELTDQVFGGCYFNIVKVSDMSIDGMP 566
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 77.4 bits (189), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 54/169 (32%), Positives = 82/169 (49%), Gaps = 15/169 (9%)
Query 8 KYCIIMCIYHAIPLLDYAISGQDGQLLVTNIEDLPIPEFDNIGMEAVPVVQLVNSSLLS- 66
++ ++MCIY +P + Y + D + + D PEF+N+GM Q +NSS +S
Sbjct 395 EHGVLMCIYSLVPQIQYDCTRLDPMVDKLDRFDYFTPEFENLGM------QPLNSSYISS 448
Query 67 --TNIGKYSFFGYNPRYYNWKTKIDRVHGAFTTDLKDWVAPIDDSYLQQWLPLTSSQPSL 124
T K GY PRY +KT +D HG F D ++ S ++W P L
Sbjct 449 FCTTDPKNPVLGYQPRYSEYKTALDVNHGQFAQ--SDALSSWSVSRFRRWTTF----PQL 502
Query 125 LYPFFKVNPNTLDNIFAVKADSTWATDQLLVNCNIDCKVVRPLSQDGIP 173
FK++P L++IF V + T A D + CN + V +S DG+P
Sbjct 503 EIADFKIDPGCLNSIFPVDYNGTEANDCVYGGCNFNIVKVSDMSVDGMP 551
Lambda K H a alpha
0.321 0.139 0.440 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 439831946280