bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-24_CDS_annotation_glimmer3.pl_2_1
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
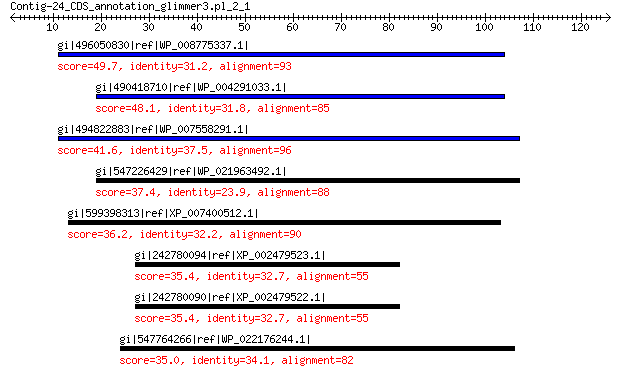
gi|496050830|ref|WP_008775337.1| hypothetical protein 49.7 2e-05
gi|490418710|ref|WP_004291033.1| hypothetical protein 48.1 7e-05
gi|494822883|ref|WP_007558291.1| hypothetical protein 41.6 0.017
gi|547226429|ref|WP_021963492.1| putative uncharacterized protein 37.4 0.82
gi|599398313|ref|XP_007400512.1| hypothetical protein PHACADRAFT... 36.2 3.4
gi|242780094|ref|XP_002479523.1| C6 transcription factor, putative 35.4 6.0
gi|242780090|ref|XP_002479522.1| C6 transcription factor, putative 35.4 6.5
gi|547764266|ref|WP_022176244.1| aAA ATPase 35.0 9.4
>gi|496050830|ref|WP_008775337.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448894|gb|EEO54685.1| hypothetical protein BSCG_01610 [Bacteroides sp. 2_2_4]
Length=154
Score = 49.7 bits (117), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/98 (30%), Positives = 52/98 (53%), Gaps = 5/98 (5%)
Query 11 PIESLRFY---TDEDGCVHMNSDVNLLMNAERI--RNQIGEENYLNIVRSIQPSRSPYKQ 65
P++ F D D + ++SD+ +L N +R+ +Q Y N + +P + +
Sbjct 34 PVDQFLFQEVSVDGDTSIRLSSDIYMLFNQQRLDKLSQTSLLEYFNNISVTEPRFNELRS 93
Query 66 KLDDDALISTLKSRYLQSPAEIDAWMASLDQKYEDMVA 103
KL D+ LIS +KSR++QS +E+ AW L ++ +A
Sbjct 94 KLGDEQLISFVKSRFIQSKSELMAWSNYLMNSTDEQIA 131
>gi|490418710|ref|WP_004291033.1| hypothetical protein [Bacteroides eggerthii]
gi|217986637|gb|EEC52971.1| hypothetical protein BACEGG_02722 [Bacteroides eggerthii DSM
20697]
Length=155
Score = 48.1 bits (113), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 50/89 (56%), Gaps = 6/89 (7%)
Query 19 TDEDGCVHMNSDVNLLMNAERI----RNQIGEENYLNIVRSIQPSRSPYKQKLDDDALIS 74
D + + SD+ +L N +R+ R+Q+ E Y + + +P S ++K+ D+ L S
Sbjct 46 CDGKKSIRITSDIYMLFNQQRLDKLTRSQLVE--YFDNLSVSEPKMSDLRKKMTDEQLCS 103
Query 75 TLKSRYLQSPAEIDAWMASLDQKYEDMVA 103
+KSR++Q+P+E+ AW L + M+A
Sbjct 104 FVKSRFIQTPSELMAWSQYLMSSQDAMIA 132
>gi|494822883|ref|WP_007558291.1| hypothetical protein [Bacteroides plebeius]
gi|198272098|gb|EDY96367.1| hypothetical protein BACPLE_00803 [Bacteroides plebeius DSM 17135]
Length=140
Score = 41.6 bits (96), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 58/119 (49%), Gaps = 24/119 (20%)
Query 11 PIESLR-------FYTDE-------DGCVHMNSDVNLLMNAERIRNQIGEE--NYLNIVR 54
P+ SLR FY +E V +D+ +++N R+ + + +YL+ R
Sbjct 3 PLTSLRVPSPVEDFYREEVQTPLGSTPAVTYRNDIYMILNQRRLDSMTLAQFSDYLDHDR 62
Query 55 SIQPSRSPYKQKLDDDALISTLKSRYLQSPAEIDAWMASLD-------QKYEDMVAEVK 106
S S ++K+ D+ L +KSRY+Q P+E+ AW + LD QK E+ V VK
Sbjct 63 SAS-QLSQMREKMSDEQLHQFVKSRYIQHPSELRAWASYLDIEYSKEIQKLEEAVEAVK 120
>gi|547226429|ref|WP_021963492.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103381|emb|CCY83992.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=152
Score = 37.4 bits (85), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 21/91 (23%), Positives = 47/91 (52%), Gaps = 4/91 (4%)
Query 19 TDEDGCVHMNSDVNLLMNAERIRNQIGEENYLNIVRSIQP---SRSPYKQKLDDDALIST 75
+D+ + D+ +L N R+ + +G + + + P S + ++ + D+ L+
Sbjct 44 SDKKTAIAYVDDIYMLFNQNRL-DSVGRDTIQKWLDGLTPRSDSLAKLRENVTDEQLMEI 102
Query 76 LKSRYLQSPAEIDAWMASLDQKYEDMVAEVK 106
KSRY+QS +E+ AW L+ Y +++ ++
Sbjct 103 CKSRYIQSSSELLAWSEYLNANYASILSSIE 133
>gi|599398313|ref|XP_007400512.1| hypothetical protein PHACADRAFT_178009 [Phanerochaete carnosa
HHB-10118-sp]
gi|409041885|gb|EKM51370.1| hypothetical protein PHACADRAFT_178009 [Phanerochaete carnosa
HHB-10118-sp]
Length=381
Score = 36.2 bits (82), Expect = 3.4, Method: Composition-based stats.
Identities = 29/90 (32%), Positives = 43/90 (48%), Gaps = 10/90 (11%)
Query 13 ESLRFYTDEDGCVHMNSDVNLLMNAERIRNQIGEENYLNIVRSIQPSRSPYKQKLDDDAL 72
ES R +E G + D L + RI +I E N+L ++S PS +P+ DDA
Sbjct 108 ESRRQLKEELGAKTLPPDHPLTRHVHRIVTRILEANHLGHLKSSTPSTAPWSAGTHDDA- 166
Query 73 ISTLKSRYLQSPAEIDAWMASLDQKYEDMV 102
YL A++ A S D+++E MV
Sbjct 167 -------YLTPEAQLPA--GSGDKEWELMV 187
>gi|242780094|ref|XP_002479523.1| C6 transcription factor, putative [Talaromyces stipitatus ATCC
10500]
gi|218719670|gb|EED19089.1| C6 transcription factor, putative [Talaromyces stipitatus ATCC
10500]
Length=734
Score = 35.4 bits (80), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 18/55 (33%), Positives = 28/55 (51%), Gaps = 0/55 (0%)
Query 27 MNSDVNLLMNAERIRNQIGEENYLNIVRSIQPSRSPYKQKLDDDALISTLKSRYL 81
+ S N+ +A R Q+ + N+L ++ I SP+ DDDA +S L S Y
Sbjct 184 LRSQANMQGSAPSFRRQVMDVNFLCVIPPIAVPASPWTTVTDDDAFVSHLISLYF 238
>gi|242780090|ref|XP_002479522.1| C6 transcription factor, putative [Talaromyces stipitatus ATCC
10500]
gi|218719669|gb|EED19088.1| C6 transcription factor, putative [Talaromyces stipitatus ATCC
10500]
Length=738
Score = 35.4 bits (80), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 18/55 (33%), Positives = 28/55 (51%), Gaps = 0/55 (0%)
Query 27 MNSDVNLLMNAERIRNQIGEENYLNIVRSIQPSRSPYKQKLDDDALISTLKSRYL 81
+ S N+ +A R Q+ + N+L ++ I SP+ DDDA +S L S Y
Sbjct 184 LRSQANMQGSAPSFRRQVMDVNFLCVIPPIAVPASPWTTVTDDDAFVSHLISLYF 238
>gi|547764266|ref|WP_022176244.1| aAA ATPase [Firmicutes bacterium CAG:552]
gi|524372700|emb|CDB26678.1| aAA ATPase [Firmicutes bacterium CAG:552]
Length=1540
Score = 35.0 bits (79), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 28/91 (31%), Positives = 49/91 (54%), Gaps = 9/91 (10%)
Query 24 CVHMNSDVNLLMNAERIRNQIGEENY-LNIVRSIQPSR--SPYKQKLDDDALISTLKS-- 78
CV + D +++ RN G+ + +N+ R + + SPY K DD I ++KS
Sbjct 285 CVAIEKDRKEILSETVGRNGKGKPKFEVNLNRYTRDAMGLSPYPPKSDDKGNIRSIKSYL 344
Query 79 ---RYLQSPAEIDAWMASLDQKYE-DMVAEV 105
RYL + +I+ +++LD+KYE D+ A+
Sbjct 345 LGERYLFTYIDIEPDLSALDKKYEKDLTAKF 375
Lambda K H a alpha
0.314 0.129 0.355 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 430790896899