bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-22_CDS_annotation_glimmer3.pl_2_2
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
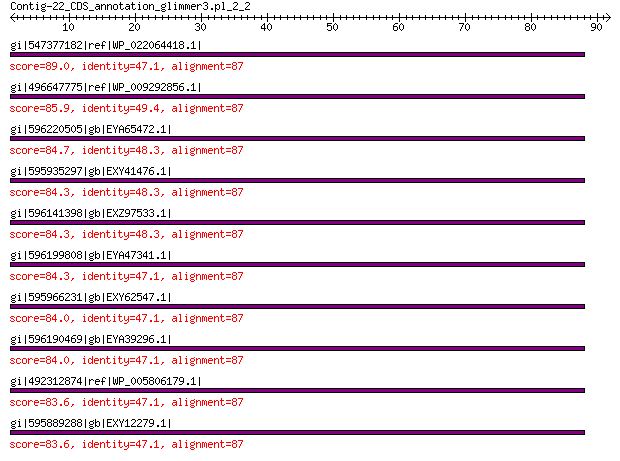
gi|547377182|ref|WP_022064418.1| uncharacterized protein 89.0 3e-20
gi|496647775|ref|WP_009292856.1| peptidase M15 85.9 5e-19
gi|596220505|gb|EYA65472.1| peptidase M15 family protein 84.7 1e-18
gi|595935297|gb|EXY41476.1| peptidase M15 family protein 84.3 1e-18
gi|596141398|gb|EXZ97533.1| peptidase M15 family protein 84.3 1e-18
gi|596199808|gb|EYA47341.1| peptidase M15 family protein 84.3 2e-18
gi|595966231|gb|EXY62547.1| peptidase M15 family protein 84.0 2e-18
gi|596190469|gb|EYA39296.1| peptidase M15 family protein 84.0 2e-18
gi|492312874|ref|WP_005806179.1| peptidase M15 83.6 3e-18
gi|595889288|gb|EXY12279.1| peptidase M15 family protein 83.6 3e-18
>gi|547377182|ref|WP_022064418.1| uncharacterized protein [Alistipes sp. CAG:157]
gi|524233222|emb|CCZ98771.1| uncharacterized protein BN505_01734 [Alistipes sp. CAG:157]
Length=132
Score = 89.0 bits (219), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 41/87 (47%), Positives = 59/87 (68%), Gaps = 0/87 (0%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
LR+A+G P+ +NSGYRS +N AVGG S+H +G AADI+V S + + LFAL++
Sbjct 44 LREAWGGPVAVNSGYRSPALNAAVGGAPRSRHMYGEAADITVGSPSANAALFALLESGGF 103
Query 61 PFDQVIYYRKAGFIHVSWSPTYRKQII 87
FDQ+I R ++HVSW T R+Q++
Sbjct 104 DFDQLIDERGYAWLHVSWCRTNRRQVL 130
>gi|496647775|ref|WP_009292856.1| peptidase M15 [Bacteroides sp. 2_1_56FAA]
gi|335942346|gb|EGN04192.1| hypothetical protein HMPREF1018_03647 [Bacteroides sp. 2_1_56FAA]
Length=131
Score = 85.9 bits (211), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 43/88 (49%), Positives = 63/88 (72%), Gaps = 2/88 (2%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
LR+ +GKPI I+SGYRS +N +V G ++SQH+ G AADI+V S++ +LF +IQ +EL
Sbjct 43 LREKYGKPIRISSGYRSAILNRSVNGATSSQHRVGEAADITVGSKEENRKLFEIIQ-LEL 101
Query 61 PFDQVIYYRKAGFIHVSWSP-TYRKQII 87
PFDQ+I R ++HVS+ RKQ++
Sbjct 102 PFDQLIDERDFSWVHVSFREGRNRKQVL 129
>gi|596220505|gb|EYA65472.1| peptidase M15 family protein [Bacteroides fragilis str. S23L24]
gi|596220788|gb|EYA65743.1| peptidase M15 family protein [Bacteroides fragilis str. S23L24]
gi|596221291|gb|EYA66190.1| peptidase M15 family protein [Bacteroides fragilis str. S23L24]
gi|598867492|gb|EYE41306.1| peptidase M15 family protein [Bacteroides fragilis str. S23L17]
Length=118
Score = 84.7 bits (208), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/88 (48%), Positives = 63/88 (72%), Gaps = 2/88 (2%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
LR+ +GKPI I+SGYRS +N +V G ++SQH+ G AADI+V S++ +LF +I ++EL
Sbjct 30 LREKYGKPIRISSGYRSAILNRSVNGATSSQHRVGEAADITVGSKEENRKLFEII-RLEL 88
Query 61 PFDQVIYYRKAGFIHVSWSP-TYRKQII 87
PFDQ+I R ++HVS+ RKQ++
Sbjct 89 PFDQLIDERDFSWVHVSFREGRNRKQVL 116
>gi|595935297|gb|EXY41476.1| peptidase M15 family protein [Bacteroides fragilis str. 3774
T13]
Length=131
Score = 84.3 bits (207), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/88 (48%), Positives = 63/88 (72%), Gaps = 2/88 (2%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
LR+ +GKPI I+SGYRS +N +V G ++SQH+ G AADI+V S++ +LF +I ++EL
Sbjct 43 LREKYGKPIRISSGYRSAILNRSVNGATSSQHRVGEAADITVGSKEENRKLFEII-RLEL 101
Query 61 PFDQVIYYRKAGFIHVSWSP-TYRKQII 87
PFDQ+I R ++HVS+ RKQ++
Sbjct 102 PFDQLIDERDFSWVHVSFREGRNRKQVL 129
>gi|596141398|gb|EXZ97533.1| peptidase M15 family protein [Bacteroides fragilis str. S23 R14]
gi|596142085|gb|EXZ97831.1| peptidase M15 family protein [Bacteroides fragilis str. S23 R14]
gi|596142457|gb|EXZ97951.1| peptidase M15 family protein [Bacteroides fragilis str. S23 R14]
gi|596145855|gb|EYA00672.1| peptidase M15 family protein [Bacteroides fragilis str. S23 R14]
gi|598870229|gb|EYE43692.1| peptidase M15 family protein [Bacteroides fragilis str. S23L17]
Length=131
Score = 84.3 bits (207), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/88 (48%), Positives = 63/88 (72%), Gaps = 2/88 (2%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
LR+ +GKPI I+SGYRS +N +V G ++SQH+ G AADI+V S++ +LF +I ++EL
Sbjct 43 LREKYGKPIRISSGYRSAILNRSVNGATSSQHRVGEAADITVGSKEENRKLFEII-RLEL 101
Query 61 PFDQVIYYRKAGFIHVSWSP-TYRKQII 87
PFDQ+I R ++HVS+ RKQ++
Sbjct 102 PFDQLIDERDFSWVHVSFREGRNRKQVL 129
>gi|596199808|gb|EYA47341.1| peptidase M15 family protein [Bacteroides fragilis str. 3719
T6]
Length=131
Score = 84.3 bits (207), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 41/88 (47%), Positives = 63/88 (72%), Gaps = 2/88 (2%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
LR+ +GKPI ++SGYRS +N +V G ++SQH+ G AADI+V S++ +LF +I ++EL
Sbjct 43 LREKYGKPIRVSSGYRSAILNRSVNGATSSQHRIGEAADITVGSKEENRKLFEII-RLEL 101
Query 61 PFDQVIYYRKAGFIHVSWSP-TYRKQII 87
PFDQ+I R ++HVS+ RKQ++
Sbjct 102 PFDQLIDERDFSWVHVSFREGRNRKQVL 129
>gi|595966231|gb|EXY62547.1| peptidase M15 family protein [Bacteroides fragilis str. 3986
N(B)19]
Length=131
Score = 84.0 bits (206), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 41/88 (47%), Positives = 63/88 (72%), Gaps = 2/88 (2%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
LR+ +GKPI ++SGYRS +N +V G ++SQH+ G AADI+V S++ +LF +I ++EL
Sbjct 43 LREKYGKPIRLSSGYRSAILNRSVNGATSSQHRIGEAADITVGSKEENRKLFEII-RLEL 101
Query 61 PFDQVIYYRKAGFIHVSWSP-TYRKQII 87
PFDQ+I R ++HVS+ RKQ++
Sbjct 102 PFDQLIDERDFSWVHVSFREGRNRKQVL 129
>gi|596190469|gb|EYA39296.1| peptidase M15 family protein [Bacteroides fragilis str. 20793-3]
Length=131
Score = 84.0 bits (206), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 41/88 (47%), Positives = 63/88 (72%), Gaps = 2/88 (2%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
LR+ +GKPI I+SGYRS +N +V G ++SQH+ G AADI+V S++ +LF +I ++EL
Sbjct 43 LREKYGKPIRISSGYRSAILNRSVNGATSSQHRLGEAADITVGSKEENRKLFEII-RLEL 101
Query 61 PFDQVIYYRKAGFIHVSWSP-TYRKQII 87
PFDQ+I + ++HVS+ RKQ++
Sbjct 102 PFDQLIDEKDFSWVHVSFREGRNRKQVL 129
>gi|492312874|ref|WP_005806179.1| peptidase M15 [Bacteroides fragilis]
gi|404587187|gb|EKA91737.1| hypothetical protein HMPREF1203_00569 [Bacteroides fragilis HMW
610]
gi|596099911|gb|EXZ63837.1| peptidase M15 family protein [Bacteroides fragilis str. 3725
D9(v)]
gi|663464423|gb|KER59296.1| peptidase M15 [Bacteroides fragilis]
Length=131
Score = 83.6 bits (205), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 41/88 (47%), Positives = 63/88 (72%), Gaps = 2/88 (2%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
LR+ +GKPI ++SGYRS +N +V G ++SQH+ G AADI+V S++ LF +I+K EL
Sbjct 43 LREKYGKPIRVSSGYRSAVLNRSVNGATSSQHRLGQAADITVGSKEGNRRLFEIIRK-EL 101
Query 61 PFDQVIYYRKAGFIHVSW-SPTYRKQII 87
PFDQ+I + ++HVS+ + RKQ++
Sbjct 102 PFDQLIDEKDFSWVHVSFRTGKNRKQVL 129
>gi|595889288|gb|EXY12279.1| peptidase M15 family protein [Bacteroides fragilis str. 1007-1-F
#8]
gi|596049346|gb|EXZ22268.1| peptidase M15 family protein [Bacteroides fragilis str. S13 L11]
gi|596064598|gb|EXZ32934.1| peptidase M15 family protein [Bacteroides fragilis str. 1007-1-F
#4]
gi|596071131|gb|EXZ38484.1| peptidase M15 family protein [Bacteroides fragilis str. 1007-1-F
#7]
gi|596161504|gb|EYA13967.1| peptidase M15 family protein [Bacteroides fragilis str. 1007-1-F
#10]
gi|596166226|gb|EYA18179.1| peptidase M15 family protein [Bacteroides fragilis str. 1007-1-F
#3]
gi|596172898|gb|EYA23855.1| peptidase M15 family protein [Bacteroides fragilis str. 1007-1-F
#9]
gi|598879125|gb|EYE52075.1| peptidase M15 family protein [Bacteroides fragilis str. 1007-1-F
#5]
gi|598890662|gb|EYE62806.1| peptidase M15 family protein [Bacteroides fragilis str. 1007-1-F
#6]
Length=131
Score = 83.6 bits (205), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 41/88 (47%), Positives = 62/88 (70%), Gaps = 2/88 (2%)
Query 1 LRDAFGKPICINSGYRSNFVNEAVGGVSNSQHKFGYAADISVSSRDSISELFALIQKMEL 60
LR+ GKPI ++SGYRS +N +V G ++SQH+ G AADI+V S++ +LF +I ++EL
Sbjct 43 LREKHGKPIRVSSGYRSAILNRSVNGATSSQHRLGEAADITVGSKEENRKLFEII-RLEL 101
Query 61 PFDQVIYYRKAGFIHVSWSP-TYRKQII 87
PFDQ+I R ++HVS+ RKQ++
Sbjct 102 PFDQLIDERDFSWVHVSFREGRNRKQVL 129
Lambda K H a alpha
0.323 0.138 0.408 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 437208516180