bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-21_CDS_annotation_glimmer3.pl_2_1
Length=82
Score E
Sequences producing significant alignments: (Bits) Value
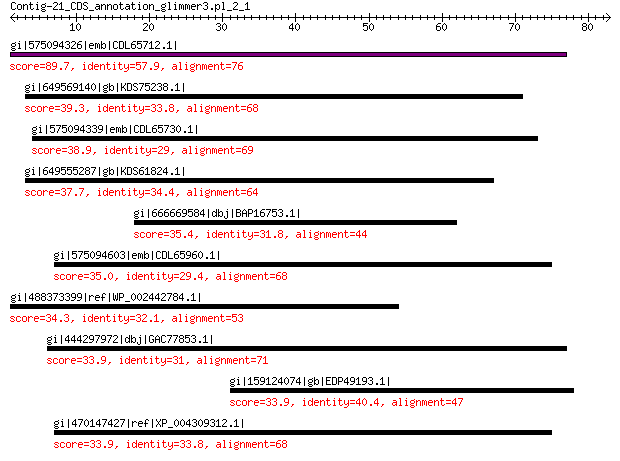
gi|575094326|emb|CDL65712.1| unnamed protein product 89.7 9e-19
gi|649569140|gb|KDS75238.1| capsid family protein 39.3 0.16
gi|575094339|emb|CDL65730.1| unnamed protein product 38.9 0.25
gi|649555287|gb|KDS61824.1| capsid family protein 37.7 0.51
gi|666669584|dbj|BAP16753.1| P0 protein 35.4 1.9
gi|575094603|emb|CDL65960.1| unnamed protein product 35.0 3.6
gi|488373399|ref|WP_002442784.1| oligoendopeptidase F 34.3 6.0
gi|444297972|dbj|GAC77853.1| major capsid protein 33.9 7.9
gi|159124074|gb|EDP49193.1| hypothetical protein AFUB_086410 33.9 8.5
gi|470147427|ref|XP_004309312.1| PREDICTED: capsid protein VP1-like 33.9 9.4
>gi|575094326|emb|CDL65712.1| unnamed protein product [uncultured bacterium]
Length=758
Score = 89.7 bits (221), Expect = 9e-19, Method: Composition-based stats.
Identities = 44/77 (57%), Positives = 54/77 (70%), Gaps = 2/77 (3%)
Query 1 LLAYRFRAYEAVYNAYYRDIRNNPFVVNGRPVYNKWLATMKGGADK-TPYELHQCNWERD 59
L AY FRAYEA+YNAY R+ RNNPFV+NG+ YN+W+ T GG+D TP +L NW+ D
Sbjct 360 LSAYPFRAYEAIYNAYIRNTRNNPFVLNGKKTYNRWITTDAGGSDTLTPRDLRFANWQSD 419
Query 60 FLTTAVPNPQQSANAPL 76
TTA+ PQQ APL
Sbjct 420 AYTTALTAPQQGV-APL 435
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 39.3 bits (90), Expect = 0.16, Method: Composition-based stats.
Identities = 23/73 (32%), Positives = 38/73 (52%), Gaps = 15/73 (21%)
Query 3 AYRFRAYEAVYNAYYRDIRNNPFVVNGRPVYNKWLATMKGGADKTP-----YELHQCNWE 57
A FRAY +YN YYRD + + ++ T+ G + P ++LH+ WE
Sbjct 9 ALPFRAYHLIYNEYYRD----------QNLTSELEITLDSGNYQLPVNSSLWQLHRRAWE 58
Query 58 RDFLTTAVPNPQQ 70
+D+ T+A+P Q+
Sbjct 59 KDYFTSALPWVQR 71
>gi|575094339|emb|CDL65730.1| unnamed protein product [uncultured bacterium]
Length=588
Score = 38.9 bits (89), Expect = 0.25, Method: Composition-based stats.
Identities = 20/69 (29%), Positives = 35/69 (51%), Gaps = 3/69 (4%)
Query 4 YRFRAYEAVYNAYYRDIRNNPFVVNGRPVYNKWLATMKGGADKTPYELHQCNWERDFLTT 63
+ F AY+ ++N +YR + + YN A + D + +ELH W++D+ T
Sbjct 180 FLFTAYQKIFNDFYR---LDDYTSVQHKSYNVDYAQGQPITDNSMFELHYRPWKKDYFTN 236
Query 64 AVPNPQQSA 72
+PNP S+
Sbjct 237 VIPNPYFSS 245
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 37.7 bits (86), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 22/69 (32%), Positives = 36/69 (52%), Gaps = 15/69 (22%)
Query 3 AYRFRAYEAVYNAYYRDIRNNPFVVNGRPVYNKWLATMKGGADKTP-----YELHQCNWE 57
A FRAY +YN YYRD + + ++ T+ G + P ++LH+ WE
Sbjct 160 ALPFRAYHLIYNEYYRD----------QNLTSELEITLDSGNYQLPVNSSLWQLHRRAWE 209
Query 58 RDFLTTAVP 66
+D+ T+A+P
Sbjct 210 KDYFTSALP 218
>gi|666669584|dbj|BAP16753.1| P0 protein [Beet western yellows virus]
Length=238
Score = 35.4 bits (80), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 14/44 (32%), Positives = 27/44 (61%), Gaps = 0/44 (0%)
Query 18 RDIRNNPFVVNGRPVYNKWLATMKGGADKTPYELHQCNWERDFL 61
R I++ + G+ ++K+L+ GA++ ++H+ N ERDFL
Sbjct 120 RIIKHQDLLARGKREFDKFLSVWCAGAERNLSQIHKTNIERDFL 163
>gi|575094603|emb|CDL65960.1| unnamed protein product [uncultured bacterium]
Length=507
Score = 35.0 bits (79), Expect = 3.6, Method: Composition-based stats.
Identities = 20/69 (29%), Positives = 32/69 (46%), Gaps = 11/69 (16%)
Query 7 RAYEAVYNAYYRDIRNNPFVVNGRPVYNKWLATMKGGADKT-PYELHQCNWERDFLTTAV 65
RAY +YN Y+RD + + + + + G D T P L + +W +D+ T A
Sbjct 128 RAYALIYNEYFRD----------QDIDEELPISFESGQDTTTPSVLQRADWRKDYFTLAR 177
Query 66 PNPQQSANA 74
P Q+ A
Sbjct 178 PFEQKGPAA 186
>gi|488373399|ref|WP_002442784.1| oligoendopeptidase F [Staphylococcus caprae]
gi|242348647|gb|EES40249.1| oligoendopeptidase F [Staphylococcus caprae M23864:W1]
Length=602
Score = 34.3 bits (77), Expect = 6.0, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 27/53 (51%), Gaps = 5/53 (9%)
Query 1 LLAYRFRAYEAVYNAYYRDIRNNPFVVNGRPVYNKWLATMKGGADKTPYELHQ 53
L +Y + A + + I+N G+P ++WL T+K G K+P EL Q
Sbjct 519 LYSYTYSAGLTIGTVVSQKIKNE-----GQPAVDQWLETLKAGGSKSPVELAQ 566
>gi|444297972|dbj|GAC77853.1| major capsid protein [uncultured marine virus]
Length=529
Score = 33.9 bits (76), Expect = 7.9, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 35/73 (48%), Gaps = 13/73 (18%)
Query 6 FRAYEAVYNAYYR--DIRNNPFVVNGRPVYNKWLATMKGGADKTPYELHQCNWERDFLTT 63
FRAY ++N ++R D++N+ V T G D T Y L + D+ T+
Sbjct 151 FRAYNLIWNEWFRSEDLQNSIVV-----------DTDNGPDDITDYVLKERGKRHDYFTS 199
Query 64 AVPNPQQSANAPL 76
A+P PQ+ + L
Sbjct 200 ALPWPQKGTSVSL 212
>gi|159124074|gb|EDP49193.1| hypothetical protein AFUB_086410 [Aspergillus fumigatus A1163]
Length=490
Score = 33.9 bits (76), Expect = 8.5, Method: Composition-based stats.
Identities = 19/49 (39%), Positives = 27/49 (55%), Gaps = 2/49 (4%)
Query 31 PVYNKWLATMKG-GADK-TPYELHQCNWERDFLTTAVPNPQQSANAPLC 77
P+ K L M+G GAD PY+ Q +W DFL + +P+Q N +C
Sbjct 108 PLEEKLLRWMEGLGADVLRPYKFVQGSWRPDFLLESSEDPEQIENYRIC 156
>gi|470147427|ref|XP_004309312.1| PREDICTED: capsid protein VP1-like, partial [Fragaria vesca subsp.
vesca]
Length=421
Score = 33.9 bits (76), Expect = 9.4, Method: Composition-based stats.
Identities = 23/73 (32%), Positives = 34/73 (47%), Gaps = 17/73 (23%)
Query 7 RAYEAVYNAYYRD--IRNNPFVVNGRPVYNKWLATMKGGADKTP---YELHQCNWERDFL 61
RAY +YN ++RD ++N+ V G G D TP Y L + D+
Sbjct 181 RAYNLIYNQWFRDENLQNSVVVDKG------------DGPDTTPSTNYTLLRRGKRHDYF 228
Query 62 TTAVPNPQQSANA 74
T+A+P PQ+ A
Sbjct 229 TSALPWPQKGGTA 241
Lambda K H a alpha
0.323 0.136 0.468 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 434005500390