bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-18_CDS_annotation_glimmer3.pl_2_7
Length=639
Score E
Sequences producing significant alignments: (Bits) Value
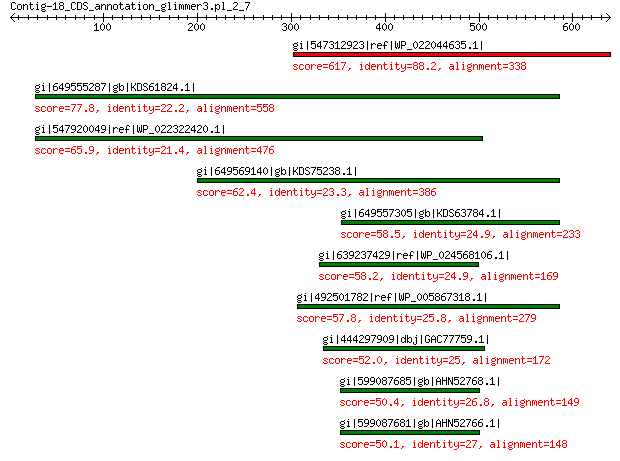
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 617 0.0
gi|649555287|gb|KDS61824.1| capsid family protein 77.8 9e-12
gi|547920049|ref|WP_022322420.1| capsid protein VP1 65.9 5e-08
gi|649569140|gb|KDS75238.1| capsid family protein 62.4 5e-07
gi|649557305|gb|KDS63784.1| capsid family protein 58.5 2e-06
gi|639237429|ref|WP_024568106.1| hypothetical protein 58.2 1e-05
gi|492501782|ref|WP_005867318.1| hypothetical protein 57.8 2e-05
gi|444297909|dbj|GAC77759.1| major capsid protein 52.0 5e-04
gi|599087685|gb|AHN52768.1| major capsid protein 50.4 0.001
gi|599087681|gb|AHN52766.1| major capsid protein 50.1 0.001
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 617 bits (1590), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 298/338 (88%), Positives = 307/338 (91%), Gaps = 0/338 (0%)
Query 302 MACAGIQGYGGLLSVPYSPDLFSNIIKQGSSPAVEIEVMNALDASTDTGFSVAVPELRLK 361
MACAGIQGYGGLLSVPYSPDLF NIIKQGSSPAVEIEVMNALD + TGFSVAVPELRL+
Sbjct 1 MACAGIQGYGGLLSVPYSPDLFGNIIKQGSSPAVEIEVMNALDLNISTGFSVAVPELRLR 60
Query 362 TKLQNWMDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSA 421
TK+QNWMDRLFVSGGRVGDVFRTLWG KSSA YVNKPDFLGVWQASINPSNVRAMANGSA
Sbjct 61 TKIQNWMDRLFVSGGRVGDVFRTLWGTKSSAIYVNKPDFLGVWQASINPSNVRAMANGSA 120
Query 422 SGEDANLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQGLHPDLASIS 481
SGEDANLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLI MLVPEPAY QGLHPDLASIS
Sbjct 121 SGEDANLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLITMLVPEPAYSQGLHPDLASIS 180
Query 482 FGDDFNPELNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTGVTIDPNMASVGE 541
FGDDFNPELNGIGFQ VPRHRF+MMPRG + QE +PW G+ GTGV +DPNM SVGE
Sbjct 181 FGDDFNPELNGIGFQLVPRHRFSMMPRGFNFTGLDQEASPWFGHTGTGVLVDPNMVSVGE 240
Query 542 EVAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGFYGDAESYSTYINPLDW 601
EVAWSWLRTDY RLHGDFAQNGNYQYWVL R FT+YFPD GTGFY D E TYINPLDW
Sbjct 241 EVAWSWLRTDYSRLHGDFAQNGNYQYWVLTRRFTTYFPDDGTGFYQDGEYTGTYINPLDW 300
Query 602 QYVFVDQTLMAGNFAYYGYFDLKVTSSLSANYMPYLGR 639
QYVFVDQTLMAGNFAYYG FDL VTSSLSANYMPYLGR
Sbjct 301 QYVFVDQTLMAGNFAYYGTFDLNVTSSLSANYMPYLGR 338
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 77.8 bits (190), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 124/570 (22%), Positives = 214/570 (38%), Gaps = 109/570 (19%)
Query 28 TIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGFKLITIATFTPDSAIYGWM 87
T++ G +P+ + + G++ R + L++ P++ P+M+ + T F P+ I+
Sbjct 27 TVNAGELIPIMCKPVVPGDKFRVNTEMLVRLAPLVAPMMHRVDVFTHYFFVPNRLIWN-- 84
Query 88 RNGRRYEPDEYTKFGKVYFPLAGPNIGNYIDPAFQIARRVRRLTFGLELNADRAKIYESW 147
++E D TK + P F T+ D A + S+
Sbjct 85 ----KWE-DFITK-----------GVDGTDSPVFP--------TYSFPSTVDTANAHNSF 120
Query 148 VSDELNSTGDDGQPTHIGRGGLWDWLGIAPGAVCPNLGIKLPSVAGVRGQFYPPSFKFNG 207
G G LWD+LG+ ++ S GV+ P FK +
Sbjct 121 -----------------GDGSLWDYLGLPSINQIGEAVFQVQSPNGVKA---PAGFKVSA 160
Query 208 APFFAYFL--SHYYYIANMQEKYMYFTRGVGEMQRVRPDGQQESLYRPFFSDVFTSFKPD 265
PF AY L + YY N+ + + T G Q + R + D FTS P
Sbjct 161 LPFRAYHLIYNEYYRDQNLTSE-LEITLDSGNYQLPVNSSLWQLHRRAWEKDYFTSALP- 218
Query 266 DFLLYLDTLSYRTRVGAEIDALTLQLEG---IPINCVQAMACAGIQGYGGLLSVPYSPDL 322
+ G E+ T+ + G IP+ + A I + + S L
Sbjct 219 -----------WVQRGPEV---TVPINGGGEIPVEMKEGFAAQKITTFPDRKPISGSEVL 264
Query 323 FS--NIIKQGSSPAVEIEVMNALD---ASTDTGFSVAVPELRLKTKLQNWMDRLFVSGGR 377
+S +++ G +++ + + D +TD V + ++R LQ W +R SG R
Sbjct 265 YSAPSVLSYGQIGSIKGQALIEPDNFVVNTDQ-MGVNINDIRTSNALQRWFERNARSGSR 323
Query 378 VGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLG--QLAACV 435
+ + +GV+SS + +P FLG + I+ S V ++ ++ AN+ ++A V
Sbjct 324 YIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAGHGISAGV 383
Query 436 DRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQGLHPDLASISFGDDFNPELNGIGF 495
+ G Y +E G M I + P Y QG+ D D + PE +G
Sbjct 384 NH--------GFTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGE 435
Query 496 QQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTGVTIDPNMASVGEEVAWSWLRTDYPRL 555
Q++ +L + N + G ++ + +
Sbjct 436 QEIKNEEL------------------YLNESDAA-----NEGTFGYTPRYAEYKYSQNEV 472
Query 556 HGDFAQNGNYQYWVLARPFTSYFPDAGTGF 585
HGDF GN +W L R F P+ T F
Sbjct 473 HGDF--RGNMAFWHLNRIFKEK-PNLNTTF 499
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 65.9 bits (159), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 102/486 (21%), Positives = 193/486 (40%), Gaps = 67/486 (14%)
Query 28 TIHPGLAVPVHHRHLNVGERIRGRIDSLLQSQPMLGPLMNGFKLITIATFTPDSAIYGWM 87
T++ G VP+ + G++ R + +SL++ P++ P+M+ + T F P+ ++
Sbjct 27 TLNMGELVPIMCMPVVSGDKFRVKTESLVRLAPLVAPMMHRVNVFTHYFFVPNRLVW--- 83
Query 88 RNGRRYEPDEYTKFGKVYFPLAGPNIGNYIDPAFQIARRVRRLTFGLELNADRAKIYESW 147
+E+ F + + G ++ P F +++N D + +
Sbjct 84 --------NEWEDF--ITKGVDGEDM-----PMFP----------KIQINQDSHLVSSAS 118
Query 148 VSDELNSTGDDGQPTHIGRGGLWDWLGIAPGAVCPNLGIKLPSVAGVRGQFYPPSFKFNG 207
+ E + G LWD+LG+ + C N + V G P F+ +
Sbjct 119 LIKE-----------YFGDSSLWDYLGLPTLSACGNKSYDV-----VNGVKVPSGFQVSA 162
Query 208 APFFAYFL--SHYYYIANMQEKYMYFTRGVGEMQRVRPDGQQESLYR-PFFSDVFTSFKP 264
PF AY L + YY N+ E + FT G G SL R + D FTS P
Sbjct 163 LPFRAYQLIYNEYYRDQNLTEP-IDFTLGSGTTVGGDQLMALMSLRRRAWEKDYFTSALP 221
Query 265 DDFLLYLDTLSYRTR-VGAEIDALTLQLEGIPINCVQAMACAGIQGYGGLLSVPYSPD-- 321
+L ++ + G +D + + + V + G+ +++ + D
Sbjct 222 --WLQRGPEVTVPVQGAGGSMDVV-YERQSDSQKWVDSSGREFENGHAYDITMARANDPN 278
Query 322 --LFSNIIKQGSSPAVEIEVMNALDASTDTGFSVAVPELRLKTKLQNWMDRLFVSGGRVG 379
L + ++ A E++ L + D + + +LR LQ W +R G R
Sbjct 279 SALMVAVNGGTNNRAPELDPNGTLKVNVDE-MGININDLRTSNALQRWFERNARGGSRYI 337
Query 380 DVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLG--QLAACVDR 437
+ + +GV+SS + +P FLG + I+ S V ++ + AN+ ++A ++
Sbjct 338 EQILSHFGVRSSDARLQRPQFLGGGRMPISVSEVLQTSSTDETSPQANMAGHGISAGIN- 396
Query 438 YCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQGLHPDLASISFGDDFNPELNGIGFQQ 497
+G +Y +E G + I + P Y QG+ D D + PE + Q+
Sbjct 397 -------NGFKHYFEEHGYIIGIMSITPRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQE 449
Query 498 VPRHRF 503
+
Sbjct 450 IKNQEL 455
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 62.4 bits (150), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 90/398 (23%), Positives = 151/398 (38%), Gaps = 63/398 (16%)
Query 200 PPSFKFNGAPFFAYFL--SHYYYIANMQEKYMYFTRGVGEMQRVRPDGQQESLYRPFFSD 257
P FK + PF AY L + YY N+ + + T G Q + R + D
Sbjct 2 PAGFKVSALPFRAYHLIYNEYYRDQNLTSE-LEITLDSGNYQLPVNSSLWQLHRRAWEKD 60
Query 258 VFTSFKPDDFLLYLDTLSYRTRVGAEIDALTLQLEG---IPINCVQAMACAGIQGYGGLL 314
FTS P + G E+ T+ + G IP+ + A I +
Sbjct 61 YFTSALP------------WVQRGPEV---TVPINGGGEIPVEMKEGFAAQKITTFPDRK 105
Query 315 SVPYSPDLFS--NIIKQGSSPAVEIEVMNALD---ASTDTGFSVAVPELRLKTKLQNWMD 369
+ S L+S +++ G +++ + + D +TD V + ++R LQ W +
Sbjct 106 PISGSEVLYSAPSVLSYGQIGSIKGQALIEPDNFVVNTDQ-MGVNINDIRTSNALQRWFE 164
Query 370 RLFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLG 429
R SG R + + +GV+SS + +P FLG + I+ S V ++ ++ AN+
Sbjct 165 RNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMA 224
Query 430 --QLAACVDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQGLHPDLASISFGDDFN 487
++A V+ G Y +E G M I + P Y QG+ D D +
Sbjct 225 GHGISAGVNH--------GFTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYF 276
Query 488 PELNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTGVTIDPNMASVGEEVAWSW 547
PE +G Q++ + + N + G ++
Sbjct 277 PEFAHLGEQEIKNEELYL-----------------------NESDAANEGTFGYTPRYAE 313
Query 548 LRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGF 585
+ +HGDF GN +W L R F P+ T F
Sbjct 314 YKYSQNEVHGDF--RGNMAFWHLNRIFKEK-PNLNTTF 348
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 58.5 bits (140), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 58/235 (25%), Positives = 93/235 (40%), Gaps = 36/235 (15%)
Query 353 VAVPELRLKTKLQNWMDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPSN 412
V + ++R LQ W +R SG R + + +GV+SS + +P FLG + I+ S
Sbjct 3 VNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSE 62
Query 413 VRAMANGSASGEDANLG--QLAACVDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYF 470
V ++ ++ AN+ ++A V+ G Y +E G M I + P Y
Sbjct 63 VLQTSSTDSTSPQANMAGHGISAGVNH--------GFTRYFEEHGYIMGIMSIRPRTGYQ 114
Query 471 QGLHPDLASISFGDDFNPELNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYAGTGV 530
QG+ D D + PE +G Q++ KN L +
Sbjct 115 QGVPKDFRKFDNMDFYFPEFAHLGEQEI--------------------KNEELYLNESDA 154
Query 531 TIDPNMASVGEEVAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGF 585
N + G ++ + +HGDF GN +W L R F P+ T F
Sbjct 155 A---NEGTFGYTPRYAEYKYSQNEVHGDF--RGNMAFWHLNRIFKEK-PNLNTTF 203
>gi|639237429|ref|WP_024568106.1| hypothetical protein [Elizabethkingia anophelis]
Length=546
Score = 58.2 bits (139), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 42/169 (25%), Positives = 76/169 (45%), Gaps = 6/169 (4%)
Query 330 GSSPAVEIEVMNALDASTDTGFSVAVPELRLKTKLQNWMDRLFVSGGRVGDVFRTLWGVK 389
G+S V I+ + L T + +LR KLQ W+++ +G R + + +GVK
Sbjct 275 GNSNPVNIDNSSNLGVDLKTASGSTINDLRRAFKLQEWLEKNARAGSRYAESILSFFGVK 334
Query 390 SSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLGQLAACVDRYCDFSGHSGIDY 449
+S + +P+FLG + I S V ++ ++ N+ V + FS
Sbjct 335 TSDGRLQRPEFLGGNKTPILISEVLQQSSTDSTTPQGNMAGHGISVGKEGGFSK------ 388
Query 450 YAKEPGTFMLIAMLVPEPAYFQGLHPDLASISFGDDFNPELNGIGFQQV 498
+ +E G + + ++P+ +Y QG+ + D F P+ IG Q V
Sbjct 389 FFEEHGYVIGLMSVIPKTSYSQGIPRHFSKFDKFDYFWPQFEHIGEQPV 437
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 57.8 bits (138), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 72/299 (24%), Positives = 117/299 (39%), Gaps = 54/299 (18%)
Query 307 IQGYGGLLSVPYSPDLFSNIIKQ---GSSPAVEIEVMN-ALDASTDTG------------ 350
IQG GG L V D ++ + + PA ++++ AL A G
Sbjct 232 IQGSGGNLDVTLKNDAHADTYRMPGTSNRPAGAMQLVGGALIAGGTDGAYLEPDNFQVNV 291
Query 351 --FSVAVPELRLKTKLQNWMDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASI 408
V++ +LR LQ W +R SG R + + +GV+SS + +P FLG + I
Sbjct 292 DELGVSINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPI 351
Query 409 NPSNVRAMANGSASGEDANLG--QLAACVDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPE 466
+ S V + ++ AN+ ++A V+ G Y +E G + I + P
Sbjct 352 SVSEVLQTSATDSTSPQANMAGHGISAGVNH--------GFKRYFEEHGYIIGIMSIRPR 403
Query 467 PAYFQGLHPDLASISFGDDFNPELNGIGFQQVPRHRFTMMPRGMDDITAHQEKNPWLGYA 526
Y QG+ D D + PE +G Q++ +++ Q
Sbjct 404 TGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKN----------EEVYLQQ--------- 444
Query 527 GTGVTIDPNMASVGEEVAWSWLRTDYPRLHGDFAQNGNYQYWVLARPFTSYFPDAGTGF 585
T N + G ++ + +HGDF GN +W L R F S P+ T F
Sbjct 445 ----TPASNNGTFGYTPRYAEYKYSMNEVHGDF--RGNMAFWHLNRIF-SESPNLNTTF 496
>gi|444297909|dbj|GAC77759.1| major capsid protein, partial [uncultured marine virus]
Length=257
Score = 52.0 bits (123), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 43/176 (24%), Positives = 75/176 (43%), Gaps = 12/176 (7%)
Query 334 AVEIEVMNALDASTDTGFSVAVPELRLKTKLQNWMDRLFVSGGRVGDVFRTLWGVKSSAP 393
+V IE + +D +V +LRL ++Q W++ +G R + WGV+SS
Sbjct 26 SVRIENLEGTQQISDADLTV--NDLRLAIRIQEWLEVNARAGSRYVEHLLAHWGVRSSDA 83
Query 394 YVNKPDFLGVWQASINPSNVRAMANGSASGE----DANLGQLAACVDRYCDFSGHSGIDY 449
+++P++LG + + S V + A + E AN+ V G +
Sbjct 84 RLDRPEYLGGGKQPVLVSEVLSTAEVAIDQEISIPQANMAGHGISV------GGSNRFKK 137
Query 450 YAKEPGTFMLIAMLVPEPAYFQGLHPDLASISFGDDFNPELNGIGFQQVPRHRFTM 505
+E G + I ++P AY QG+ + D + PE +G Q V + M
Sbjct 138 RFEEHGHILGIMSVIPRTAYQQGVDRSFSREDKFDYYFPEFAHLGEQSVNNYEVYM 193
>gi|599087685|gb|AHN52768.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=228
Score = 50.4 bits (119), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 40/149 (27%), Positives = 68/149 (46%), Gaps = 4/149 (3%)
Query 352 SVAVPELRLKTKLQNWMDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPS 411
S + ++RL + Q ++R SG R + + WGV S + + +FLG + + S
Sbjct 83 SANINDIRLAFQTQRLLERDARSGTRYVEALKAHWGVTSPDFRLQRAEFLGGGSSVVQLS 142
Query 412 NVRAMANGSASGEDANLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQ 471
V+ + + ED +G LAA SG + E G +++ + + +Y Q
Sbjct 143 PVQQQSAQTTPAEDDKVGNLAAN----GTVSGTHSWNKSFVEHGVVIILGNIRGDISYSQ 198
Query 472 GLHPDLASISFGDDFNPELNGIGFQQVPR 500
G+ + + D F P L+GIG Q VP
Sbjct 199 GIDRYWSKSTRYDFFYPVLSGIGEQSVPE 227
>gi|599087681|gb|AHN52766.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=226
Score = 50.1 bits (118), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 40/148 (27%), Positives = 68/148 (46%), Gaps = 4/148 (3%)
Query 352 SVAVPELRLKTKLQNWMDRLFVSGGRVGDVFRTLWGVKSSAPYVNKPDFLGVWQASINPS 411
S + ++RL + Q ++R SG R + + WGV S + + +FLG + + S
Sbjct 83 SANINDIRLAFQTQRLLERDARSGTRYVEALKAHWGVTSPDFRLQRAEFLGGGSSVVQLS 142
Query 412 NVRAMANGSASGEDANLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLIAMLVPEPAYFQ 471
V+ + + ED +G LAA SG + E G +++ + + +Y Q
Sbjct 143 PVQQQSAQTTPAEDDKVGNLAAN----GTVSGTHSWNKSFVEHGVVIILGNIRGDISYSQ 198
Query 472 GLHPDLASISFGDDFNPELNGIGFQQVP 499
G+ + + D F P L+GIG Q VP
Sbjct 199 GIDRYWSKSTRYDFFYPVLSGIGEQSVP 226
Lambda K H a alpha
0.321 0.140 0.442 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4823816581056