bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-11_CDS_annotation_glimmer3.pl_2_4
Length=644
Score E
Sequences producing significant alignments: (Bits) Value
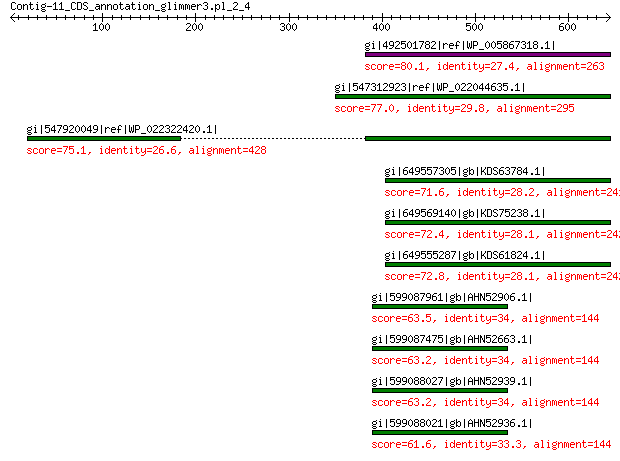
gi|492501782|ref|WP_005867318.1| hypothetical protein 80.1 2e-12
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 77.0 5e-12
gi|547920049|ref|WP_022322420.1| capsid protein VP1 75.1 6e-11
gi|649557305|gb|KDS63784.1| capsid family protein 71.6 1e-10
gi|649569140|gb|KDS75238.1| capsid family protein 72.4 3e-10
gi|649555287|gb|KDS61824.1| capsid family protein 72.8 4e-10
gi|599087961|gb|AHN52906.1| major capsid protein 63.5 4e-08
gi|599087475|gb|AHN52663.1| major capsid protein 63.2 5e-08
gi|599088027|gb|AHN52939.1| major capsid protein 63.2 6e-08
gi|599088021|gb|AHN52936.1| major capsid protein 61.6 2e-07
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 80.1 bits (196), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 72/264 (27%), Positives = 115/264 (44%), Gaps = 15/264 (6%)
Query 382 VDVTDGKLSMDALNLAQKVYNFLNRIAISGGTYRDWLETVYTGGNYMERCETPMFEGGVS 441
V+V + +S++ L + + + R A SG Y + + + + + R + P F GG
Sbjct 289 VNVDELGVSINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGR 348
Query 442 QEIVFQEVISNSASQE-EPLGTLAGRGVTTGRQKGGHIRIKVTEPCYIMCICSITPRIDY 500
I EV+ SA+ P +AG G++ G G + E YI+ I SI PR Y
Sbjct 349 TPISVSEVLQTSATDSTSPQANMAGHGISAGVNHG--FKRYFEEHGYIIGIMSIRPRTGY 406
Query 501 GQGNTWDTYLETMDDWHKPALDGIGYQDSLNGERAWWTDHIASNGGSLTKTAAGKTVAWI 560
QG D D++ P +G Q+ + E + ASN G+ G T +
Sbjct 407 QQGVPKDFRKFDNMDFYFPEFAHLGEQE-IKNEEVYLQQTPASNNGTF-----GYTPRYA 460
Query 561 NYMTNVNRTFGNFAPEMPESFMVLNRNYSMNNNGQIEDLTTYIDPVKFNYIFADTNLDAM 620
Y ++N G+F M +F LNR +S + N TT+++ N +FA
Sbjct 461 EYKYSMNEVHGDFRGNM--AFWHLNRIFSESPNLN----TTFVECNPSNRVFATAETSDD 514
Query 621 NFWVQTKFDIKVRRLISAKQIPNL 644
+W+Q D+K RL+ P L
Sbjct 515 KYWIQLYQDVKALRLMPKYGTPML 538
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 77.0 bits (188), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 88/335 (26%), Positives = 136/335 (41%), Gaps = 49/335 (15%)
Query 350 GLCLKTYNSDVYQNWINTEWIEGVTGINEASAVDV---TDGKLSMDALNLAQKVYNFLNR 406
GL Y+ D++ N I V I +A+D+ T +++ L L K+ N+++R
Sbjct 11 GLLSVPYSPDLFGNIIKQGSSPAVE-IEVMNALDLNISTGFSVAVPELRLRTKIQNWMDR 69
Query 407 IAISGGTYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEVISNSASQEEPLGTLAGR 466
+ +SGG D T++ + P F G V+Q I+ S + G+ +G
Sbjct 70 LFVSGGRVGDVFRTLWGTKSSAIYVNKPDFLG------VWQASINPSNVRAMANGSASGE 123
Query 467 GVTTGRQKG---------GH--IRIKVTEPCYIMCICSITPRIDYGQGNTWDTYLETMDD 515
G+ GH I EP M I + P Y QG D + D
Sbjct 124 DANLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLITMLVPEPAYSQGLHPDLASISFGD 183
Query 516 WHKPALDGIGYQ----------------DSLNGERAWWTDHIASNGGSLTK---TAAGKT 556
P L+GIG+Q L+ E + W H + G L + G+
Sbjct 184 DFNPELNGIGFQLVPRHRFSMMPRGFNFTGLDQEASPWFGHTGT--GVLVDPNMVSVGEE 241
Query 557 VAWINYMTNVNRTFGNFAPEMPESFMVLNRNYSM----NNNGQIED---LTTYIDPVKFN 609
VAW T+ +R G+FA + VL R ++ + G +D TYI+P+ +
Sbjct 242 VAWSWLRTDYSRLHGDFAQNGNYQYWVLTRRFTTYFPDDGTGFYQDGEYTGTYINPLDWQ 301
Query 610 YIFADTNLDAMNFWVQTKFDIKVRRLISAKQIPNL 644
Y+F D L A NF FD+ V +SA +P L
Sbjct 302 YVFVDQTLMAGNFAYYGTFDLNVTSSLSANYMPYL 336
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 75.1 bits (183), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 72/265 (27%), Positives = 118/265 (45%), Gaps = 17/265 (6%)
Query 382 VDVTDGKLSMDALNLAQKVYNFLNRIAISGGTYRDWLETVYTGGNYMERCETPMFEGGVS 441
V+V + ++++ L + + + R A G Y + + + + + R + P F GG
Sbjct 304 VNVDEMGININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGR 363
Query 442 QEIVFQEVISNSASQE-EPLGTLAGRGVTTGRQKGGHIRIKVTEPCYIMCICSITPRIDY 500
I EV+ S++ E P +AG G++ G G + E YI+ I SITPR Y
Sbjct 364 MPISVSEVLQTSSTDETSPQANMAGHGISAGINNG--FKHYFEEHGYIIGIMSITPRSGY 421
Query 501 GQGNTWD-TYLETMDDWHKPALDGIGYQDSLNGERAWWTDHIASNGGSLTKTAAGKTVAW 559
QG D T + MD ++ P + Q+ N E + ++ A N G+ G T +
Sbjct 422 QQGVPRDFTKFDNMD-FYFPEFAHLSEQEIKNQE-LFVSEDAAYNNGTF-----GYTPRY 474
Query 560 INYMTNVNRTFGNFAPEMPESFMVLNRNYSMNNNGQIEDLTTYIDPVKFNYIFADTNLDA 619
Y + + G+F + SF LNR + N TT+++ N +FA + +
Sbjct 475 AEYKYHPSEAHGDFRGNL--SFWHLNRIFEDKPNLN----TTFVECKPSNRVFATSETED 528
Query 620 MNFWVQTKFDIKVRRLISAKQIPNL 644
FWVQ D+K RL+ P L
Sbjct 529 DKFWVQMYQDVKALRLMPKYGTPML 553
Score = 45.4 bits (106), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 42/176 (24%), Positives = 77/176 (44%), Gaps = 15/176 (9%)
Query 19 SVSLHEYNMSTHDLSTIVRNTQSPGTLVPNLCLVAQKGDTFDIDIDSNVLTHPTTGPLFG 78
S+ + + +LS + T + G LVP +C+ GD F + +S V P P+
Sbjct 7 SIRMKRPRRNAFNLSYESKLTLNMGELVPIMCMPVVSGDKFRVKTESLVRLAPLVAPMMH 66
Query 79 SFKLEHHVYTGPVRL-YNSWLHNNRTKIGLNMEKVKL-PQLKVNITTLSDTPSNEEKQWV 136
+ H + P RL +N W + G++ E + + P++++N + + ++ K++
Sbjct 67 RVNVFTHYFFVPNRLVWNEW--EDFITKGVDGEDMPMFPKIQINQDSHLVSSASLIKEY- 123
Query 137 QVNPSCLLAYLGIRGYANTPNSGAGVINK---------NALPILTYFDIFKNYYAN 183
S L YLG+ + N V+N +ALP Y I+ YY +
Sbjct 124 -FGDSSLWDYLGLPTLSACGNKSYDVVNGVKVPSGFQVSALPFRAYQLIYNEYYRD 178
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 71.6 bits (174), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 68/242 (28%), Positives = 100/242 (41%), Gaps = 15/242 (6%)
Query 404 LNRIAISGGTYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEVISNSASQE-EPLGT 462
R A SG Y + + + + + R + P F GG I EV+ S++ P
Sbjct 18 FERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQAN 77
Query 463 LAGRGVTTGRQKGGHIRIKVTEPCYIMCICSITPRIDYGQGNTWDTYLETMDDWHKPALD 522
+AG G++ G G E YIM I SI PR Y QG D D++ P
Sbjct 78 MAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFA 135
Query 523 GIGYQDSLNGERAWWTDHIASNGGSLTKTAAGKTVAWINYMTNVNRTFGNFAPEMPESFM 582
+G Q+ + E + + A+N G+ G T + Y + N G+F M +F
Sbjct 136 HLGEQE-IKNEELYLNESDAANEGTF-----GYTPRYAEYKYSQNEVHGDFRGNM--AFW 187
Query 583 VLNRNYSMNNNGQIEDLTTYIDPVKFNYIFADTNLDAMNFWVQTKFDIKVRRLISAKQIP 642
LNR + N TT+++ N +FA +WVQ DIK RL+ P
Sbjct 188 HLNRIFKEKPNLN----TTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPKYGTP 243
Query 643 NL 644
L
Sbjct 244 ML 245
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 72.4 bits (176), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 68/243 (28%), Positives = 101/243 (42%), Gaps = 15/243 (6%)
Query 403 FLNRIAISGGTYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEVISNSASQE-EPLG 461
+ R A SG Y + + + + + R + P F GG I EV+ S++ P
Sbjct 162 WFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQA 221
Query 462 TLAGRGVTTGRQKGGHIRIKVTEPCYIMCICSITPRIDYGQGNTWDTYLETMDDWHKPAL 521
+AG G++ G G E YIM I SI PR Y QG D D++ P
Sbjct 222 NMAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEF 279
Query 522 DGIGYQDSLNGERAWWTDHIASNGGSLTKTAAGKTVAWINYMTNVNRTFGNFAPEMPESF 581
+G Q+ + E + + A+N G+ G T + Y + N G+F M +F
Sbjct 280 AHLGEQE-IKNEELYLNESDAANEGTF-----GYTPRYAEYKYSQNEVHGDFRGNM--AF 331
Query 582 MVLNRNYSMNNNGQIEDLTTYIDPVKFNYIFADTNLDAMNFWVQTKFDIKVRRLISAKQI 641
LNR + N TT+++ N +FA +WVQ DIK RL+
Sbjct 332 WHLNRIFKEKPNLN----TTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPKYGT 387
Query 642 PNL 644
P L
Sbjct 388 PML 390
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 72.8 bits (177), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 68/243 (28%), Positives = 101/243 (42%), Gaps = 15/243 (6%)
Query 403 FLNRIAISGGTYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEVISNSASQE-EPLG 461
+ R A SG Y + + + + + R + P F GG I EV+ S++ P
Sbjct 313 WFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQA 372
Query 462 TLAGRGVTTGRQKGGHIRIKVTEPCYIMCICSITPRIDYGQGNTWDTYLETMDDWHKPAL 521
+AG G++ G G E YIM I SI PR Y QG D D++ P
Sbjct 373 NMAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEF 430
Query 522 DGIGYQDSLNGERAWWTDHIASNGGSLTKTAAGKTVAWINYMTNVNRTFGNFAPEMPESF 581
+G Q+ + E + + A+N G+ G T + Y + N G+F M +F
Sbjct 431 AHLGEQE-IKNEELYLNESDAANEGTF-----GYTPRYAEYKYSQNEVHGDFRGNM--AF 482
Query 582 MVLNRNYSMNNNGQIEDLTTYIDPVKFNYIFADTNLDAMNFWVQTKFDIKVRRLISAKQI 641
LNR + N TT+++ N +FA +WVQ DIK RL+
Sbjct 483 WHLNRIFKEKPNLN----TTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPKYGT 538
Query 642 PNL 644
P L
Sbjct 539 PML 541
>gi|599087961|gb|AHN52906.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=210
Score = 63.5 bits (153), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 49/144 (34%), Positives = 65/144 (45%), Gaps = 3/144 (2%)
Query 390 SMDALNLAQKVYNFLNRIAISGGTYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEV 449
+++ L A ++ L R A SG Y + ++ + G N+M+ P F GG S I V
Sbjct 68 TINQLRQAFQIQKLLERDARSGTRYSEIVKAHF-GVNFMDVTYRPEFLGGTSTPINVTSV 126
Query 450 ISNSASQEEPLGTLAGRGVTTGRQKGGHIRIKVTEPCYIMCICSITPRIDYGQGNTWDTY 509
S S P GTLA G T GG TE C +M I S+ + Y QG
Sbjct 127 PQTSESGTTPQGTLAAFGTAT--INGGGFTKSFTEHCIVMGIASVRADLTYQQGLNRMFS 184
Query 510 LETMDDWHKPALDGIGYQDSLNGE 533
T D++ PAL IG Q LN E
Sbjct 185 RSTRYDFYFPALAHIGEQSVLNKE 208
>gi|599087475|gb|AHN52663.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=210
Score = 63.2 bits (152), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 49/144 (34%), Positives = 65/144 (45%), Gaps = 3/144 (2%)
Query 390 SMDALNLAQKVYNFLNRIAISGGTYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEV 449
+++ L A ++ L R A SG Y + ++ + G N+M+ P F GG S I V
Sbjct 68 TINQLRQAFQIQKLLERDARSGTRYSEIVKAHF-GVNFMDVTYRPEFLGGTSTPINVTSV 126
Query 450 ISNSASQEEPLGTLAGRGVTTGRQKGGHIRIKVTEPCYIMCICSITPRIDYGQGNTWDTY 509
S S P GTLA G T GG TE C +M I S+ + Y QG
Sbjct 127 PQTSESGTTPQGTLAAFGTAT--INGGGFTKSFTEHCILMGIASVRADLTYQQGLNRMFS 184
Query 510 LETMDDWHKPALDGIGYQDSLNGE 533
T D++ PAL IG Q LN E
Sbjct 185 RSTRYDFYFPALAHIGEQSVLNKE 208
>gi|599088027|gb|AHN52939.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=219
Score = 63.2 bits (152), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 49/144 (34%), Positives = 65/144 (45%), Gaps = 3/144 (2%)
Query 390 SMDALNLAQKVYNFLNRIAISGGTYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEV 449
+++ L A ++ L R A SG Y + ++ + G N+M+ P F GG S I V
Sbjct 77 TINQLRQAFQIQKLLERDARSGTRYAEIVKAHF-GVNFMDVTYRPEFLGGTSTPINVTSV 135
Query 450 ISNSASQEEPLGTLAGRGVTTGRQKGGHIRIKVTEPCYIMCICSITPRIDYGQGNTWDTY 509
S S P GTLA G T GG TE C +M I S+ + Y QG
Sbjct 136 PQTSESGTTPQGTLAAFGTAT--VNGGGFTKSFTEHCIVMGIASVRADLTYQQGLNRMFS 193
Query 510 LETMDDWHKPALDGIGYQDSLNGE 533
T D++ PAL IG Q LN E
Sbjct 194 RSTRYDFYFPALAHIGEQAVLNKE 217
>gi|599088021|gb|AHN52936.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=220
Score = 61.6 bits (148), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 48/144 (33%), Positives = 65/144 (45%), Gaps = 3/144 (2%)
Query 390 SMDALNLAQKVYNFLNRIAISGGTYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEV 449
+++ L A ++ L R A SG Y + ++ + G N+M+ P F GG S + V
Sbjct 78 TINQLRQAFQIQKLLERDARSGTRYSEIVKAHF-GVNFMDVTYRPEFLGGSSTPVNVTSV 136
Query 450 ISNSASQEEPLGTLAGRGVTTGRQKGGHIRIKVTEPCYIMCICSITPRIDYGQGNTWDTY 509
S S P GTLA G T GG TE C +M I S+ + Y QG
Sbjct 137 PQTSESGTTPQGTLAAFGTAT--INGGGFTKSFTEHCIVMGIASVRADLTYQQGLNRMFS 194
Query 510 LETMDDWHKPALDGIGYQDSLNGE 533
T D++ PAL IG Q LN E
Sbjct 195 RSTRYDFYFPALAHIGEQSVLNKE 218
Lambda K H a alpha
0.315 0.133 0.397 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4873649396976