bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-10_CDS_annotation_glimmer3.pl_2_9
Length=317
Score E
Sequences producing significant alignments: (Bits) Value
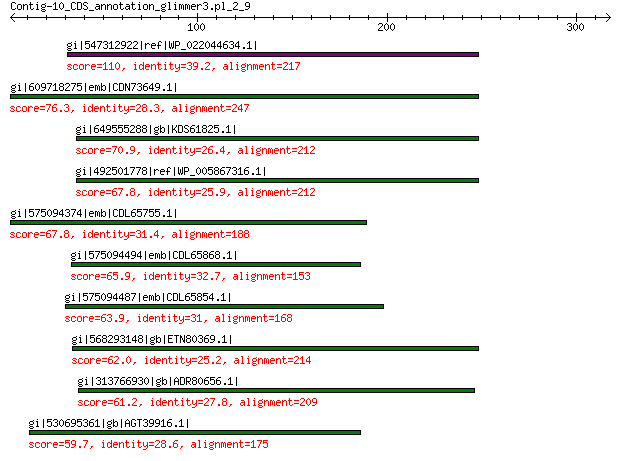
gi|547312922|ref|WP_022044634.1| putative replication initiation... 110 2e-24
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 76.3 8e-13
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 70.9 8e-11
gi|492501778|ref|WP_005867316.1| hypothetical protein 67.8 1e-09
gi|575094374|emb|CDL65755.1| unnamed protein product 67.8 2e-09
gi|575094494|emb|CDL65868.1| unnamed protein product 65.9 8e-09
gi|575094487|emb|CDL65854.1| unnamed protein product 63.9 3e-08
gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 62.0 2e-07
gi|313766930|gb|ADR80656.1| putative replication initiation protein 61.2 3e-07
gi|530695361|gb|AGT39916.1| replication initiator 59.7 6e-07
>gi|547312922|ref|WP_022044634.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
gi|524208442|emb|CCZ76638.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
Length=320
Score = 110 bits (276), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 85/236 (36%), Positives = 125/236 (53%), Gaps = 50/236 (21%)
Query 31 LLYIPAACGECYECRKKKQREYQIRLQEEIR-HTHGE--FLTLTIDNDWYkklekeipkl 87
+L +P CG C+ C+K +Y+IRL E+R + G F+TLT ++D + K
Sbjct 39 ILEVP--CGYCHSCQKSYNNQYRIRLLYELRKYPPGTCLFVTLTFNDD-------SLEKF 89
Query 88 pkDTIKETKINGEYDNLIATVALRKFLERCRKETGKSIKHWCITELGEENGRIHIHGIFF 147
KDT K A+R FL+R RK GK I+HW + E G +GR H HGI F
Sbjct 90 SKDTNK---------------AVRLFLDRFRKVYGKQIRHWFVCEFGTLHGRPHYHGILF 134
Query 148 GENINQ---------------VIQKNWKYGFIYIGQYVNERTVMYITKYMFKQCEYNKLF 192
N+ Q ++ WKYGF+++G YV++ T YITKY+ K +K+
Sbjct 135 --NVPQALIDGYDSDMPGHHPLLASCWKYGFVFVG-YVSDETCSYITKYVTKSINGDKV- 190
Query 193 KGKVLSSSGIGSGYVSRFDSSRNKYKEDGNTN-ETYQFRNGVKVNLPNYYRNKIYS 247
+ +V+SS GIGS Y++ +SS +K GN + + NG + +P YY NKI+S
Sbjct 191 RPRVISSFGIGSNYLNTEESSLHKL---GNQRYQPFMVLNGFQQAMPRYYYNKIFS 243
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 76.3 bits (186), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 70/259 (27%), Positives = 112/259 (43%), Gaps = 52/259 (20%)
Query 1 MCLYPKLIRNPKYKPTKTNGGNPPVCYDTRLLYIPAACGECYECRKKKQREYQIRLQEEI 60
MCL P ++ TNG Y T+ CG+C ECRK + + RL EE+
Sbjct 1 MCLTPVTLKKTG---ATTNG------YATQSF----PCGKCLECRKARTNSWFARLTEEL 47
Query 61 RHT-HGEFLTLTIDNDWYkklekeipklpkDTIKETKINGEYDNLIATVALRKF---LER 116
+ + F+TLT + + DN + ++ R F ++R
Sbjct 48 KVSKSAHFVTLTYSDVYLPYS---------------------DNGLISLDYRDFQLFMKR 86
Query 117 CRKETGKSIKHWCITELGEENGRIHIHGIFFG-ENINQVIQKNWKYGFIYIGQYVNERTV 175
RK IK++ + E G + R H H I FG ENI+ + + W+ G ++ G V +++
Sbjct 87 ARKLQKSKIKYFLVGEYGAQTYRPHYHAIVFGVENIDAFLGE-WRMGNVHAGT-VTAKSI 144
Query 176 MYITKYMFKQCE-------YNKLFKGKVLSSSGIGSGYVSRFDSSRNKYKEDGNTNETYQ 228
Y KY K + K L S G+G +S S KY +D + + ++
Sbjct 145 YYTLKYCTKSITEGPDKDPDDDRKPEKALMSKGLG---LSHLTESMIKYYKD-DVSRSFS 200
Query 229 FRNGVKVNLPNYYRNKIYS 247
G + LP YYR+K++S
Sbjct 201 LLGGTTIALPRYYRDKVFS 219
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 70.9 bits (172), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 56/225 (25%), Positives = 107/225 (48%), Gaps = 27/225 (12%)
Query 36 AACGECYECRKKKQREYQIRLQEEI-RHTHGEFLTLTIDNDWYkklekeipklpkDTIKE 94
CG C CRK K++ + RLQ E + F+TLT D++ + +D K
Sbjct 15 VPCGRCVNCRKNKRQSWVYRLQAEADEYPFSLFVTLTYDDE-----HIPTAMIGEDLFKT 69
Query 95 TKINGEYDNLIATVALRKFLERCRKETGK-SIKHWCITELGEENGRIHIHGIFF-----G 148
T +++ ++ F++R RK+ + ++++ +E G + GR H H I F G
Sbjct 70 TV------GVVSKRDIQLFMKRLRKKYAQYRLRYFLTSEYGSQGGRPHYHMILFGFPFTG 123
Query 149 ENINQVIQKNWKYGFIYIGQYVNERTVMYITKYMFKQCEYNKLFKGK------VLSSSGI 202
++ ++ + WK GF+ + + + Y+TKYM+++ + KG +L S
Sbjct 124 KHGGDLLAECWKNGFVQ-AHPLTTKEISYVTKYMYEKSMIPDILKGVKEYQPFMLCSKMP 182
Query 203 GSGYVSRFDSSRNKYKEDGNTNETYQFRNGVKVNLPNYYRNKIYS 247
G GY + + Y+ + + + NG+++ +P YY +K+Y
Sbjct 183 GIGYHFLREQILDFYRL--HPRDYVRAFNGMRMAMPRYYADKLYD 225
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 67.8 bits (164), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 55/225 (24%), Positives = 106/225 (47%), Gaps = 27/225 (12%)
Query 36 AACGECYECRKKKQREYQIRLQEEI-RHTHGEFLTLTIDNDWYkklekeipklpkDTIKE 94
CG C CRK K++ + RLQ E + F+TLT D++ + +D K
Sbjct 15 VPCGRCVNCRKNKRQSWVYRLQAEADEYPFSLFVTLTYDDE-----HMPTAMIGEDLFKS 69
Query 95 TKINGEYDNLIATVALRKFLERCRKETGK-SIKHWCITELGEENGRIHIHGIFF-----G 148
T +++ ++ F++R RK+ + ++++ +E G + GR H H I F G
Sbjct 70 TV------GVVSKRDIQLFMKRLRKKYDQYRLRYFLTSEYGSQGGRPHYHMILFGFPFTG 123
Query 149 ENINQVIQKNWKYGFIYIGQYVNERTVMYITKYMFKQCEYNKLFKGK------VLSSSGI 202
++ ++ + WK GF+ + + + Y+TKYM+++ + K +L S
Sbjct 124 KHGGDLLAECWKNGFVQ-AHPLTTKEIAYVTKYMYEKSMVPDILKDVKEYQPFMLCSRIP 182
Query 203 GSGYVSRFDSSRNKYKEDGNTNETYQFRNGVKVNLPNYYRNKIYS 247
G GY + + Y+ + + + NG+++ +P YY +K+Y
Sbjct 183 GIGYHFLREQILDFYR--LHPRDYVRAFNGMRMAMPRYYADKLYD 225
>gi|575094374|emb|CDL65755.1| unnamed protein product [uncultured bacterium]
Length=487
Score = 67.8 bits (164), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 59/202 (29%), Positives = 88/202 (44%), Gaps = 37/202 (18%)
Query 1 MCLYPKLIRNPKYKPTKTNGGNPPVCYDTRLLYIPAACGECYECRKKKQREYQIRLQEEI 60
MC P +IRN N Y +P CG CY+C+ K ++Q+R EE+
Sbjct 1 MCFSPIIIRN--------NSSYIHTHYTYADYVVP--CGHCYDCKSAKTTDWQVRCSEEL 50
Query 61 -RHTHGEFLTLTIDNDWYkklekeipklpkDTIKETKINGEYDNLIATVALRKFLERCRK 119
++ F TLT+D + DT T +G + ++ FL+R RK
Sbjct 51 NNNSQSYFYTLTLDPRF------------IDTYG-TLPDGSPRYVFNKRHIQLFLKRLRK 97
Query 120 ETGK---SIKHWCITELGEENGRIHIHGIFF-GENINQ-----VIQKNWKYGFIYIGQ-- 168
K S+K+ + ELGE R H H IF+ ++N +++ +W GFI G
Sbjct 98 ALSKYNISLKYVIVGELGETTHRPHYHAIFYLSSSVNPFKFRIMVRNSWSLGFIKSGDNN 157
Query 169 --YVNERTVMYITKYMFKQCEY 188
+N V Y+ KYM K Y
Sbjct 158 GIILNNDAVSYVIKYMHKTDSY 179
>gi|575094494|emb|CDL65868.1| unnamed protein product [uncultured bacterium]
Length=348
Score = 65.9 bits (159), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 50/177 (28%), Positives = 76/177 (43%), Gaps = 34/177 (19%)
Query 33 YIPAACGECYECRKKKQREYQIR-LQEEIRHTHGEFLTLTIDNDWYkklekeipklpkDT 91
YI CG+C CR R++ R + E HTH FLTLT D+D ++
Sbjct 59 YIIIPCGKCVGCRLAYSRQWADRCMLESSYHTHSYFLTLTYDDD---------NLPLSES 109
Query 92 IKETKINGEYDNLIATVALRKFLERCRK------ETGKSIKHWCITELGEENGRIHIHGI 145
I + Y+ + ++ F++R R+ + IK++C E G + R H H I
Sbjct 110 INQDTGEINYNATLVKKDIQDFIKRLRRFCEYNIDDNLHIKYFCAGEYGSQTFRPHYHMI 169
Query 146 FFGENINQV-----------------IQKNWKYGFIYIGQYVNERTVMYITKYMFKQ 185
+G IN + I K WK GF+ IG+ V T Y +Y+ K+
Sbjct 170 LYGFPINDLKLYKMSLDGYNYYNSATIDKLWKKGFVVIGE-VTWDTCAYTARYILKK 225
>gi|575094487|emb|CDL65854.1| unnamed protein product [uncultured bacterium]
Length=332
Score = 63.9 bits (154), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 52/192 (27%), Positives = 90/192 (47%), Gaps = 38/192 (20%)
Query 30 RLLYIPAACGECYECRKKKQREYQIRL-QEEIRHTHGEFLTLTIDNDWYkklekeipklp 88
RLL +P C +C CR K RE+ R+ E++ H FLTLT +++
Sbjct 47 RLLMLP--CRQCVGCRLSKSREWANRVVMEQLYHVESWFLTLTYNDE-----------HL 93
Query 89 kDTIKETKINGEYDNLIATVA---LRKFLERCRKETGKSIKHWCITELGEENGRIHIHGI 145
+ + GE ++ T+ L+KFL+R RK +G+ ++ + E G N R H H +
Sbjct 94 PRSFPVDEATGEILSVHGTLVKEDLQKFLKRLRKNSGQKLRFFAAGEYGSLNMRPHYHLL 153
Query 146 FFGENI-----------------NQVIQKNWKYGFIYIGQYVNERTVMYITKYMFKQCE- 187
FG ++ + +++K W +GF +G+ V ++ Y+ +Y K+
Sbjct 154 IFGLHLEDLQLLRKSPLGDEYYTSSLLEKCWPFGFHILGR-VTWQSAAYVARYTMKKASK 212
Query 188 -YNK-LFKGKVL 197
Y+K L+K L
Sbjct 213 GYDKDLYKKAAL 224
>gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 [Necator americanus]
Length=345
Score = 62.0 bits (149), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 54/225 (24%), Positives = 94/225 (42%), Gaps = 32/225 (14%)
Query 34 IPAACGECYECRKKKQREYQIRL-QEEIRHTHGEFLTLTIDNDWYkklekeipklpkDTI 92
+P CG C C++++ + RL QEE++H + F+TLT D + +
Sbjct 18 VPVPCGRCPPCKRRRVDSWVFRLLQEELQHENASFVTLTYDTRFVPISKNGFM------- 70
Query 93 KETKINGEYDNLIATVALRKFLERCRKET-GKSIKHWCITELGEENGRIHIHGIFFGENI 151
T GE+ ++++R RK G+ +K++ E G + R H H I FG
Sbjct 71 --TLDRGEFP---------RYMKRLRKLVPGRKLKYYMCGEYGSQRFRPHYHAIIFGVPQ 119
Query 152 NQVIQKNWKYG----FIYIGQYVNERTVMYITKYMFKQCEYNKLFKGK-----VLSSSGI 202
+ + W + V +++ Y KY+ K K + L S G+
Sbjct 120 DSLFADAWTLNGDSLGGVVVGTVTGKSIAYTMKYIDKSTWKQKHGRDDRVPEFSLMSKGM 179
Query 203 GSGYVSRFDSSRNKYKEDGNTNETYQFRNGVKVNLPNYYRNKIYS 247
G Y++ +Y ++ + G ++ +P YYR KIYS
Sbjct 180 GVSYLT---PQMVEYHKEDISRLFCTREGGSRIAMPRYYRQKIYS 221
>gi|313766930|gb|ADR80656.1| putative replication initiation protein [Uncultured Microviridae]
Length=402
Score = 61.2 bits (147), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 58/250 (23%), Positives = 103/250 (41%), Gaps = 68/250 (27%)
Query 37 ACGECYECRKKKQREYQIRLQEEIR-HTHGEFLTLTIDNDWYkklekeipklpkDTIKET 95
CG+C+ CR + RE+ IR E + H H F+TLTI+ ET
Sbjct 129 PCGQCWGCRLQHSREWAIRCMHEAQMHDHNCFITLTIN-------------------PET 169
Query 96 KINGEYDNLIATVALRKFLERCRKETGKSIKHWCITELGEENGRIHIHGIFFGEN----- 150
+ ++F+ R R++ GK IK++ E G+EN R H H I FG +
Sbjct 170 LERRPRPWSLEKKEFQEFVHRLRRKIGKKIKYFHCGEYGDENKRPHYHAIIFGYDFPDKQ 229
Query 151 -----------INQVIQKNWKYGFIYIGQYVNERTVMYITKYMFKQC------------- 186
I+ ++ W +G+ IG E + Y+ +Y+ K+
Sbjct 230 LWERKLGNELYISPELENLWPHGYHRIGACTYE-SAHYVARYVMKRAKGEGPPEQYINPE 288
Query 187 ----------EYNKLFKG-KVLSSSGIGSGYVSRFDSSRNKYKEDGNTNETYQFRNGVKV 235
+Y + +G K +GIG+ + ++ + D + ++ Y +G+K+
Sbjct 289 TGEVEYDLDNQYATMSRGNKKQPQNGIGNQWYWKYGWT------DAHCHD-YIVHDGIKM 341
Query 236 NLPNYYRNKI 245
+P YY ++
Sbjct 342 KVPRYYDKEL 351
>gi|530695361|gb|AGT39916.1| replication initiator [Marine gokushovirus]
Length=289
Score = 59.7 bits (143), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 50/195 (26%), Positives = 79/195 (41%), Gaps = 40/195 (21%)
Query 11 PKYKPTKTNGGNPPVCYDTRLLYIPA---ACGECYECRKKKQREYQIR-LQEEIRHTHGE 66
P Y P + V +D + CG+C CR R++ IR + E H
Sbjct 2 PCYHPLVAYKCDGKVVFDKPFAFARGFNLPCGQCIGCRLDYSRQWAIRCVHEAQTHEDNC 61
Query 67 FLTLTIDNDWYkklekeipklpkDTIKETKINGEYDNLIATVALRKFLERCRKETGKSIK 126
F+TLT DN+ I + K DN ++F++R RK+ I+
Sbjct 62 FITLTFDNE---------------HIAKRKNPESLDN----TEFQRFMKRLRKKYPHKIR 102
Query 127 HWCITELGEENGRIHIHGIFFGEN----------------INQVIQKNWKYGFIYIGQYV 170
+ E G++N R H H + FG + ++Q + + W YGF IG V
Sbjct 103 FFHCGEYGDQNKRPHYHALLFGHDFKDKKLWSNKGDFKLFVSQELAELWPYGFHTIGA-V 161
Query 171 NERTVMYITKYMFKQ 185
+ T Y +Y+ K+
Sbjct 162 SFDTAAYCARYVMKK 176
Lambda K H a alpha
0.319 0.138 0.431 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1760970990960