bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
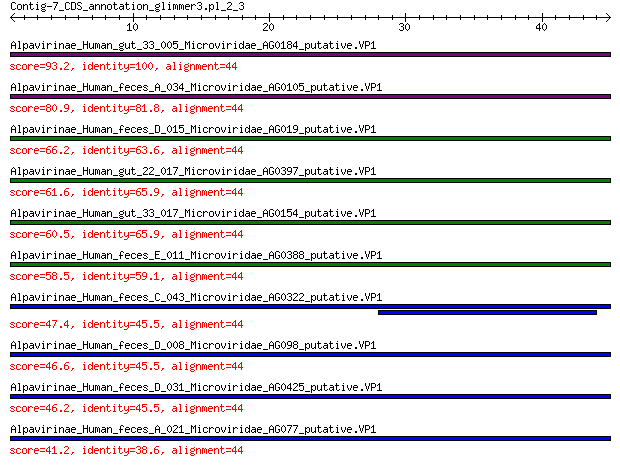
Query= Contig-7_CDS_annotation_glimmer3.pl_2_3
Length=44
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1 93.2 9e-26
Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1 80.9 2e-21
Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1 66.2 3e-16
Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1 61.6 1e-14
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 60.5 3e-14
Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1 58.5 2e-13
Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1 47.4 1e-09
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 46.6 2e-09
Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1 46.2 3e-09
Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1 41.2 1e-07
> Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1
Length=618
Score = 93.2 bits (230), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 44/44 (100%), Positives = 44/44 (100%), Gaps = 0/44 (0%)
Query 1 VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 44
VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY
Sbjct 575 VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 618
> Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1
Length=618
Score = 80.9 bits (198), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 36/44 (82%), Positives = 39/44 (89%), Gaps = 0/44 (0%)
Query 1 VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 44
VNPSV+NPIF V DGSWNTDQLL NC FDV+VARNLSYDG+PY
Sbjct 575 VNPSVLNPIFAVSVDGSWNTDQLLCNCQFDVKVARNLSYDGMPY 618
> Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1
Length=584
Score = 66.2 bits (160), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 28/44 (64%), Positives = 35/44 (80%), Gaps = 0/44 (0%)
Query 1 VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 44
VNPS+++PIF V AD W+TD L+N FD+RVARNL YDG+PY
Sbjct 541 VNPSILDPIFAVNADSYWDTDTFLINAAFDIRVARNLDYDGMPY 584
> Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1
Length=607
Score = 61.6 bits (148), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 29/44 (66%), Positives = 35/44 (80%), Gaps = 0/44 (0%)
Query 1 VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 44
VNPSV++PIFGV AD +W++DQLLVN VARNLS DG+PY
Sbjct 564 VNPSVLDPIFGVAADSTWDSDQLLVNSYIGCYVARNLSRDGVPY 607
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 60.5 bits (145), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 29/44 (66%), Positives = 34/44 (77%), Gaps = 0/44 (0%)
Query 1 VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 44
VNPSV++PIFGV AD +W+TDQLLVN V RNLS DG+PY
Sbjct 571 VNPSVLDPIFGVKADSTWDTDQLLVNSYIGCYVVRNLSRDGVPY 614
> Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1
Length=622
Score = 58.5 bits (140), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 26/44 (59%), Positives = 32/44 (73%), Gaps = 0/44 (0%)
Query 1 VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 44
VNP+ ++ IF VVAD W TDQLL+NCD +V R LS DG+PY
Sbjct 579 VNPNTLDSIFAVVADSIWETDQLLINCDVSCKVVRPLSQDGMPY 622
> Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1
Length=602
Score = 47.4 bits (111), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 20/44 (45%), Positives = 30/44 (68%), Gaps = 0/44 (0%)
Query 1 VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 44
V P V++PIF +W++DQ LVN F+V+ +NL Y+G+PY
Sbjct 559 VTPRVLDPIFVQECTDTWDSDQFLVNVSFNVKPVQNLDYNGMPY 602
Score = 20.4 bits (41), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 7/16 (44%), Positives = 13/16 (81%), Gaps = 0/16 (0%)
Query 28 DFDVRVARNLSYDGLP 43
DF++ A+N+S++ LP
Sbjct 182 DFNLLAAKNVSFNVLP 197
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 46.6 bits (109), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 20/44 (45%), Positives = 27/44 (61%), Gaps = 0/44 (0%)
Query 1 VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 44
VNP V++ IF V D + +TDQ L D++ RN YDG+PY
Sbjct 575 VNPRVLDSIFNVKCDSTIDTDQFLTTLYMDIKAVRNFDYDGMPY 618
> Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1
Length=614
Score = 46.2 bits (108), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 20/44 (45%), Positives = 27/44 (61%), Gaps = 0/44 (0%)
Query 1 VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 44
VNP V++ IF V D + +TDQ L D++ RN YDG+PY
Sbjct 571 VNPRVLDSIFNVKCDSTIDTDQFLTALYMDIKAVRNFDYDGMPY 614
> Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1
Length=622
Score = 41.2 bits (95), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 17/44 (39%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 1 VNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 44
VNP++++ IF V AD + +TD ++ RNL Y G+PY
Sbjct 579 VNPAILDDIFAVKADSTMDTDTFKTRMMMSIKCVRNLDYSGMPY 622
Lambda K H a alpha
0.321 0.141 0.452 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3716982