bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
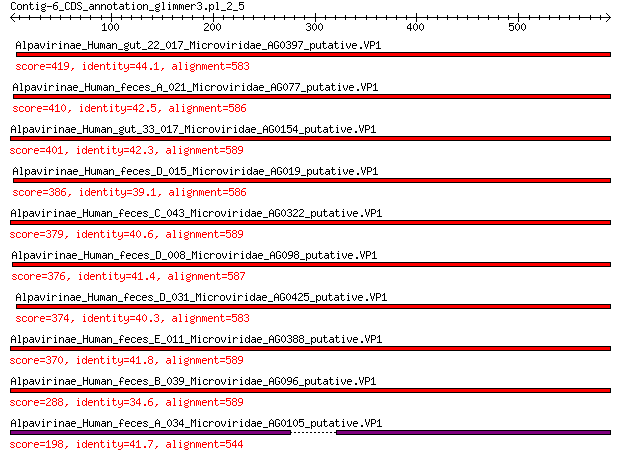
Query= Contig-6_CDS_annotation_glimmer3.pl_2_5
Length=589
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1 419 2e-140
Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1 410 8e-137
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 401 3e-133
Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1 386 1e-127
Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1 379 1e-124
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 376 2e-123
Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1 374 1e-122
Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1 370 4e-121
Alpavirinae_Human_feces_B_039_Microviridae_AG096_putative.VP1 288 3e-90
Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1 198 7e-57
> Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1
Length=607
Score = 419 bits (1078), Expect = 2e-140, Method: Compositional matrix adjust.
Identities = 257/619 (42%), Positives = 355/619 (57%), Gaps = 48/619 (8%)
Query 7 LKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEV-LPGDTFKIDLKSFTRTQPLNTAA 65
+ +L+N R+GFD+ K FTAK GELLPV W++ +PGD +K +++ FTRTQP+ T+A
Sbjct 1 MSNLQNHPHRSGFDIGRKNAFTAKVGELLPVY-WDISMPGDKYKFNVEYFTRTQPVETSA 59
Query 66 FARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTDLSSIS 125
+ R+REY+DFY VP LLW A + LTQM D Q S N+ L P LS +
Sbjct 60 YTRLREYFDFYAVPLRLLWKSAPSVLTQMQDINQLQALSFTQNL-SLGSYFPSLTLSRFT 118
Query 126 RYLNSLASNSTAVTNKA---NYFGYNRALCSAKLMECLGYGNLYYYAESTSNTFAKRPLH 182
LN L S N + N FG++RA + KL LGYGN++ S+SN + L
Sbjct 119 AVLNRLNGGSNVPGNSSTFLNEFGFSRADLAFKLFSYLGYGNVWSSEFSSSNRWWSTSLK 178
Query 183 YNLN----------VSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSSGMML 232
+ V+LF LLAYQKIY D++R SQWE +PS +NVDY TG S +++
Sbjct 179 GGGSYTQQYVQDSYVNLFPLLAYQKIYQDFFRWSQWENSNPSSYNVDYF---TGVSPLLV 235
Query 233 SF---NYSDFYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASI----SMSVPVVAGSSA 285
S + ++++ +MFDL+YCNW KD+ GV+PN Q+GDVA + S VV G
Sbjct 236 SSLPEASNAYWKSPTMFDLKYCNWNKDMLMGVLPNSQFGDVAVLDIDNSGKPDVVLGLGN 295
Query 286 AlinsritssnstttlrFP-------TDPAIPDATPLLT-----HPSFSILALRQAEFLQ 333
A + S S+ T P + +P + L F++LALRQAE LQ
Sbjct 296 ANSTVGVASYVSSNTASIPFFALKASSANTLPVGSTLRVDLASLKSQFTVLALRQAEALQ 355
Query 334 KWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINEVVNQNITG-SNAA 392
+WKEI+QSG+ DY+EQ+ KH+ V+ S MCTY+GGIS +LDI+EVVN N+ + A
Sbjct 356 RWKEISQSGDSDYREQIRKHFGVNLPQALSNMCTYIGGISRNLDISEVVNNNLAAEGDTA 415
Query 393 DIAGKGTGVSNGVINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIPEFD 452
IAGKG G NG + + + VVMCIYH +PL+DYT + +A IPEFD
Sbjct 416 VIAGKGVGAGNGSFTYTTN-EHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFD 474
Query 453 RVGMQTVPLSYVSNGPLSVLPLSIPN--EIGYAPRYIDYKTDIDTSIGAFKTSLKNWVIS 510
+GM+T+P++ + N P + SI N GY PRY ++KT +D GAF T+LK+WV
Sbjct 475 NIGMETLPMTQIFNSPKA----SIVNLFNAGYNPRYFNWKTKLDVINGAFTTTLKSWVSP 530
Query 511 YDNQSLANQFGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEADSSIDTDQFLC 570
L+ FG+ E V + NY FKVNP+ L+P+F V ADS+ D+DQ L
Sbjct 531 VTESLLSGWFGFGYS--EGDVNSQNKVVLNYKFFKVNPSVLDPIFGVAADSTWDSDQLLV 588
Query 571 STFFDVKVVRNLDTDGLPY 589
+++ V RNL DG+PY
Sbjct 589 NSYIGCYVARNLSRDGVPY 607
> Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1
Length=622
Score = 410 bits (1055), Expect = 8e-137, Method: Compositional matrix adjust.
Identities = 249/630 (40%), Positives = 354/630 (56%), Gaps = 54/630 (9%)
Query 4 IMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQPLNT 63
+ S L+N R+GFDL+ + FT+KAGELLPV +PGD + I FTRTQP++T
Sbjct 3 LFSYDDLKNHPRRSGFDLTKRICFTSKAGELLPVYWKPTIPGDKWNISTNWFTRTQPVDT 62
Query 64 AAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTDLSS 123
AA+ R++EY D++FVP +L+ +++TQM DNP A+ + + D MP+T L S
Sbjct 63 AAYTRVKEYVDWFFVPLNLIQKGIESAITQMVDNPVSAMSAIENRAITTD--MPYTTLLS 120
Query 124 ISRYLNSLASNS--TAVTNKANYFGYNRALCSAKLMECLGYGNLYYYAESTSNT------ 175
+SR L L S + K N FG++RA SAKL++ L YGN S +T
Sbjct 121 LSRALYMLNGKSYVNSHAGKLNMFGFSRADLSAKLLQMLKYGNFINPEHSGLDTPMFGYS 180
Query 176 ---FAKRPLHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSSGMML 232
A+ +N V++ + YQKIY+DY+R QWE+ P +N DY Y+ G
Sbjct 181 TVKLAQFSYLWNQAVNVLPIFCYQKIYSDYFRFQQWEKPVPYTYNADY--YAGGDIFPNP 238
Query 233 SFNYSDFYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASISMSV--------PVVAGSS 284
+ ++ NY+ FDLRYCNW +DL+ G +PNQQ+G+V+ + M+V PV G++
Sbjct 239 TTMPDAYWNNYTPFDLRYCNWNRDLYTGFLPNQQFGNVSVVDMTVSSDEIMAAPVTFGAT 298
Query 285 A------AlinsritssnstttlrFPTDPAIPDATPLLTH-------PSFSILALRQAEF 331
+ TSS T+ P+ +P T L +FSILALRQAEF
Sbjct 299 GNRVAVTKAALASSTSSTGIGTVSTPSGGTVPVGTSLYAQVQQRDLTGAFSILALRQAEF 358
Query 332 LQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINEVVNQNIT---G 388
LQKWKEI SG++DY+ Q+EKH+ V S M Y+GG + +DI+EVVNQN+T G
Sbjct 359 LQKWKEIALSGDQDYRSQIEKHFGVKLPAELSYMSQYIGGQFAQMDISEVVNQNLTDQAG 418
Query 389 SNAAD----IAGKGTGVSNGVINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAA 444
S+AA IAGKG +G +N+ ++ ++G++M IYH +PL+DY + +A
Sbjct 419 SDAAQYPALIAGKGVNSGDGNVNYTAR-QHGIIMGIYHAVPLLDYERTGQDQDLLITSAE 477
Query 445 DFAIPEFDRVGMQTVPLSYVSN-----GPLSVLPLSIPNEIGYAPRYIDYKTDIDTSIGA 499
++AIPEFD +GMQT+PL + N G IGY PRY+++KTDID GA
Sbjct 478 EWAIPEFDAIGMQTLPLGTLFNSNKVSGDSDFRLHGAAYPIGYVPRYVNWKTDIDEIFGA 537
Query 500 FKTSLKNWVISYDNQSLANQFGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEA 559
F++S K WV D + N + + S +NY FKVNP L+ +FAV+A
Sbjct 538 FRSSEKTWVAPIDADFITNWVKNVADNASA-----VQSLFNYNWFKVNPAILDDIFAVKA 592
Query 560 DSSIDTDQFLCSTFFDVKVVRNLDTDGLPY 589
DS++DTD F +K VRNLD G+PY
Sbjct 593 DSTMDTDTFKTRMMMSIKCVRNLDYSGMPY 622
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 401 bits (1030), Expect = 3e-133, Method: Compositional matrix adjust.
Identities = 249/624 (40%), Positives = 351/624 (56%), Gaps = 45/624 (7%)
Query 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEV-LPGDTFKIDLKSFTRTQ 59
MA L +L+N R+GFD+ K FTAK GELLPV W++ +PGD ++ +++ FTRTQ
Sbjct 1 MAFYTGLSNLQNHPHRSGFDIGRKNAFTAKVGELLPVY-WDISMPGDKYRFNVEYFTRTQ 59
Query 60 PLNTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYD-NPQHALD-SSPLNVVKLDGSMP 117
P+ T+A+ R+REY+DFY VP LLW A + LTQM D N AL + L++ S+
Sbjct 60 PVATSAYTRLREYFDFYAVPLRLLWKSAPSVLTQMQDVNQIQALSLTQNLSLGTYLPSLT 119
Query 118 FTDLSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYYYAESTSNTFA 177
L RYLN + N FG++RA S KL+ LGYGNL S N +
Sbjct 120 IGTLGWAIRYLNGNTWEPETASYLRNAFGFSRADLSFKLLSYLGYGNLIETPPSLGNRWW 179
Query 178 KRPLH-------------YNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYS 224
L N V++F LL YQKIY D++R QWE+ +PS +NVDY S
Sbjct 180 STSLKNTDDGANYTQQYIQNTIVNIFPLLTYQKIYQDFFRWPQWEKSNPSSYNVDYFSGS 239
Query 225 TGSSGMMLSFNYSDFYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASISMSVP----VV 280
+ S L S ++++ +MFDL+YCNW KD+ GV+PN Q+GDVA + +S VV
Sbjct 240 SPSIVSDLPAASSAYWKSDTMFDLKYCNWNKDMLMGVLPNSQFGDVAVLDISSSGDSDVV 299
Query 281 AGSSAAlinsritssnstttlrFP--------TDPAIPDATPLLTHPS----FSILALRQ 328
G I S+ ++ + P ++P + L S F++LALRQ
Sbjct 300 LGVDPHKSTLGIGSAITSKSAVVPLFALDASTSNPVSVGSKLHLDLSSIKSQFNVLALRQ 359
Query 329 AEFLQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINEVVNQNITG 388
AE LQ+WKEI+QSG+ DY+EQ+ KH+ V S +CTY+GGIS +LDI+EVVN N+
Sbjct 360 AEALQRWKEISQSGDSDYREQILKHFGVKLPQALSNLCTYIGGISRNLDISEVVNNNLAA 419
Query 389 -SNAADIAGKGTGVSNGVINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFA 447
+ A IAGKG G NG + + + V+M IYH +PL+DYT + +A
Sbjct 420 EEDTAVIAGKGVGTGNGSFTYTTN-EHCVIMGIYHAVPLLDYTLTGQDGQLLVTDAESLP 478
Query 448 IPEFDRVGMQTVPLSYVSNGPLSVLPLSIPNEIGYAPRYIDYKTDIDTSIGAFKTSLKNW 507
IPEFD +G++ +P++ + N L+ ++ N GY PRY ++KT +D GAF T+LK+W
Sbjct 479 IPEFDNIGLEVLPMAQIFNSSLAT-AFNLFNA-GYNPRYFNWKTKLDVINGAFTTTLKSW 536
Query 508 VISYDNQSLAN--QFGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEADSSIDT 565
V L+ +FG S + NY FKVNP+ L+P+F V+ADS+ DT
Sbjct 537 VSPVSESLLSGWARFG------ASDSKTGTKAVLNYKFFKVNPSVLDPIFGVKADSTWDT 590
Query 566 DQFLCSTFFDVKVVRNLDTDGLPY 589
DQ L +++ VVRNL DG+PY
Sbjct 591 DQLLVNSYIGCYVVRNLSRDGVPY 614
> Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1
Length=584
Score = 386 bits (991), Expect = 1e-127, Method: Compositional matrix adjust.
Identities = 229/610 (38%), Positives = 341/610 (56%), Gaps = 52/610 (9%)
Query 4 IMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQPLNT 63
+ +L S++N+ R+GFDLSSK F+AK GELLP+K +PGD F + + FTRTQP+NT
Sbjct 3 LFNLSSVKNRPRRSGFDLSSKVAFSAKVGELLPIKWTLTMPGDKFSLKEQHFTRTQPVNT 62
Query 64 AAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTDLSS 123
+A+ R+REYYD+++ P LLW A + Q+ N QHA SS V L +MP
Sbjct 63 SAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQHA--SSFDGSVLLGSNMPCFSADQ 120
Query 124 ISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNL-----------YYYAEST 172
IS+ L+ + S K NYFG+NRA + KL++ L YGN+ Y +
Sbjct 121 ISQSLDMMKS-------KLNYFGFNRADLAYKLIQYLRYGNVRTGVGTSGSRNYGTSVDV 173
Query 173 SNTFAKRPLHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSSGMML 232
++ + +N +S+F +LAY+K DY+R +QW+ +P +N+DY Y + +L
Sbjct 174 KDSSYNQNRAFNHALSVFPILAYKKFCQDYFRLTQWQVSAPYLWNIDY--YDGKGATTIL 231
Query 233 SFNYSD---FYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASISMS-----VPVVAGSS 284
+ S ++E+ + FDL YCNW KD+F G +P+ QYGD + + +S PV+ +
Sbjct 232 PADLSKSVTYFEHDTFFDLEYCNWNKDMFFGSLPDAQYGDTSVVDISYGTTGAPVITAQN 291
Query 285 AAlinsritssnstttlrFPTDPAIPDATPLLTHPSFSILALRQAEFLQKWKEITQSGNK 344
+ T+ + +D T+ + +LALR+ E LQ+++EI+
Sbjct 292 LQSPVNSSTAIGT-------SDKFSTQLIEAGTNLTLDVLALRRGEALQRFREISLCTPL 344
Query 345 DYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINEVVNQNITGSNAADIAGKGTGVSNG 404
+Y+ Q++ H+ V G S M TY+GG +SSLDI+EVVN NIT SN A IAGKG G G
Sbjct 345 NYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNTNITESNEALIAGKGIGTGQG 404
Query 405 VINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIPEFDRVGMQTVPLSYV 464
F ++ +GV+MCIYH +PL+DY P + F +PE D +G++ + ++Y
Sbjct 405 NEEFYAK-DWGVLMCIYHSVPLLDYVISAPDPQLFASMNTSFPVPELDAIGLEPITVAYY 463
Query 465 SNGPLSV-----LPLSIPNEIGYAPRYIDYKTDIDTSIGAFKTSLKNWVISYDNQSLANQ 519
SN P+ + + + +GY PRY +KT ID +GAF T+ K WV + +N
Sbjct 464 SNNPIELPSTGGITDAPTTTVGYLPRYYAWKTSIDYVLGAFTTTEKEWVAPITPELWSNM 523
Query 520 FGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEADSSIDTDQFLCSTFFDVKVV 579
P+ + NY FKVNP+ L+P+FAV ADS DTD FL + FD++V
Sbjct 524 L--------KPL-GTKGTGINYNFFKVNPSILDPIFAVNADSYWDTDTFLINAAFDIRVA 574
Query 580 RNLDTDGLPY 589
RNLD DG+PY
Sbjct 575 RNLDYDGMPY 584
> Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1
Length=602
Score = 379 bits (972), Expect = 1e-124, Method: Compositional matrix adjust.
Identities = 239/623 (38%), Positives = 350/623 (56%), Gaps = 55/623 (9%)
Query 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQP 60
MAN+ S S++NK +R+GFD S +FTAKAGELLPV +LPG ++L SFTRT P
Sbjct 1 MANLFSYGSVKNKPARSGFDWSEHFSFTAKAGELLPVYWKMLLPGTKVNLNLSSFTRTMP 60
Query 61 LNTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTD 120
+NTAA+ R++EYYD+YFVP L+ +L QM D P A V LD +P+T+
Sbjct 61 VNTAAYTRVKEYYDWYFVPLRLINKSIGQALVQMQDQPVQATSIVANKSVTLD--LPWTN 118
Query 121 LSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYYYAESTS------- 173
+++ LN + +TNK N G+++A SAKL+ L YGN YY ++ +
Sbjct 119 AATMFTLLNYA---NVILTNKYNLDGFSKAATSAKLLRYLRYGNCYYTSDPSKVGKNKNF 175
Query 174 --------NTFAKRPLHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYST 225
N A + + +N+ L AYQKIY D++R QWE P +N DY Y+
Sbjct 176 GLSSKDDFNLLAAKNVSFNV----LPLAAYQKIYCDWFRFEQWENACPYTYNFDY--YNG 229
Query 226 GSSGMMLSFNYSDFYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASIS--------MSV 277
G+ ++ N +F+ N ++ LRY N+ KDLF GV+P+ Q+G VA+++ +S
Sbjct 230 GNVFAGVTANPENFWSNDNILSLRYANYNKDLFMGVMPSSQFGSVATVNVANFSSSNLSS 289
Query 278 PVVAGSSAAlinsritssnstttlrFPT-DPAIPDATPLLTH-------PSFSILALRQA 329
P+ S+ ++ + +S+ + + + D + D +LT SF IL+ R A
Sbjct 290 PLRNLSNVGMVTASNNASSGSPLILRSSQDTSAGDGFGILTSNIFSTLSASFDILSFRIA 349
Query 330 EFLQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINEVVNQNITGS 389
E QKWKE+TQ + YKEQ+E H+NV + S+ C Y+GG SS + I+EV+N N+ S
Sbjct 350 EATQKWKEVTQCAKQGYKEQLEAHFNVKLSEALSDHCRYIGGTSSGVTISEVLNTNLE-S 408
Query 390 NAADIAGKGTGVSNGVINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIP 449
AADI GKG G S G F + +G++MCIYH P++DY + A D IP
Sbjct 409 AAADIKGKGVGGSFGSETFETN-EHGILMCIYHATPVLDYLRSGQDLQLLSTLATDIPIP 467
Query 450 EFDRVGMQTVPLSYVSN---GPLSVLPLSIPNEIGYAPRYIDYKTDIDTSIGAFKTSLKN 506
EFD +GM+ +P+ + N + L SIP +GY+PRYI YKT +D GAF+T+L +
Sbjct 468 EFDHIGMEALPIETLFNEQSTEATALINSIP-VLGYSPRYIAYKTSVDWVSGAFETTLDS 526
Query 507 WVISYDNQSLANQFGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEADSSIDTD 566
WV ++ Q + P+S + S NY FKV P L+P+F E + D+D
Sbjct 527 WVAPL---TVNEQITKLLFNPDS----GSVYSMNYGFFKVTPRVLDPIFVQECTDTWDSD 579
Query 567 QFLCSTFFDVKVVRNLDTDGLPY 589
QFL + F+VK V+NLD +G+PY
Sbjct 580 QFLVNVSFNVKPVQNLDYNGMPY 602
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 376 bits (966), Expect = 2e-123, Method: Compositional matrix adjust.
Identities = 243/630 (39%), Positives = 345/630 (55%), Gaps = 56/630 (9%)
Query 3 NIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVL-PGDTFKIDLKSFTRTQPL 61
++ ++ +++N R+GFDLS++ FT+KAGELLPV W+++ PGD+FKI + FTRTQPL
Sbjct 2 SLFNMSAVKNHPRRSGFDLSNRVCFTSKAGELLPV-FWDIVYPGDSFKIKTQLFTRTQPL 60
Query 62 NTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGS-MPFTD 120
NTAA+ R+REY DFYFVP L+ T+LTQM DNP A S VV D +P
Sbjct 61 NTAAYTRIREYLDFYFVPLRLINKNLPTALTQMQDNPVQATGLSSNKVVTTDIPWVPVHS 120
Query 121 LSSISRYLNSLA--SNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYYYAESTSNTFAK 178
S L A S + ++ N+ G++ SAKL+ L YGN + + +
Sbjct 121 SSGSYSSLTGFADVRGSVSSSDIENFLGFDSITQSAKLLMYLRYGN--FLSSVVPDEQKS 178
Query 179 RPLHYNLN-------------VSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYST 225
L +L+ V + L YQKIYAD++R +QWE+ P +N D+ YS
Sbjct 179 IGLSSSLDLRNSETVSTGYTSVHILPLATYQKIYADFFRFTQWEKNQPYTYNFDW--YSG 236
Query 226 GSSGMMLSFNY--SDFYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASISMS-----VP 278
G+ L+ +Y + ++F LRY NW KD+F GV+P+ Q GDV+ + S P
Sbjct 237 GNVLASLTTQALAKKYYSDDNLFTLRYANWPKDMFMGVMPDSQLGDVSIVDTSGSEGTFP 296
Query 279 VV------AGSSAAlinsritssnstttlrFPTDPAIPDATPLLTHP--------SFSIL 324
V GS A L+ TS + ++L T A+ +T + SFSIL
Sbjct 297 VGLYNFADGGSRAGLVAVSGTSPAAGSSLDMQTTSALSASTKYGVYAQQVAGLGSSFSIL 356
Query 325 ALRQAEFLQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINEVVNQ 384
LR AE +QK++E++Q ++D + Q+ H+ VS S+ C YLGG SS++D++EVVN
Sbjct 357 QLRMAEAVQKYREVSQFADQDARGQIMAHFGVSLSPVLSDKCMYLGGSSSNIDLSEVVNT 416
Query 385 NITGSNAADIAGKGTGVSNGVI--NFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVN 442
NITG N A+IAGKG G G NF++ YG++M IYH +PL+DY ++ N
Sbjct 417 NITGDNIAEIAGKGVGTGQGSFSGNFDT---YGIIMGIYHNVPLLDYVITGQPQNLLYTN 473
Query 443 AADFAIPEFDRVGMQTVPLSYVSNGPLSVLPLSI---PNEIGYAPRYIDYKTDIDTSIGA 499
AD PE+D +GMQT+ N + +GY PR+ D KT D +GA
Sbjct 474 TADLPFPEYDSIGMQTIQFGRFVNSKAVGWTSGVDYRTQTMGYLPRFFDVKTRYDEVLGA 533
Query 500 FKTSLKNWVISYDNQSLANQFGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEA 559
F+++LKNWV D +L SV + N NY FKVNP L+ +F V+
Sbjct 534 FRSTLKNWVAPLDPANLPQWLQTSVTSSGKLFLN-----LNYGFFKVNPRVLDSIFNVKC 588
Query 560 DSSIDTDQFLCSTFFDVKVVRNLDTDGLPY 589
DS+IDTDQFL + + D+K VRN D DG+PY
Sbjct 589 DSTIDTDQFLTTLYMDIKAVRNFDYDGMPY 618
> Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1
Length=614
Score = 374 bits (960), Expect = 1e-122, Method: Compositional matrix adjust.
Identities = 235/629 (37%), Positives = 338/629 (54%), Gaps = 61/629 (10%)
Query 7 LKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVL-PGDTFKIDLKSFTRTQPLNTAA 65
+ +++N R+GFDLS++ FT+KAGELLPV W+++ PGD+FKI + FTRTQPLNTAA
Sbjct 1 MSAVKNHPRRSGFDLSNRVCFTSKAGELLPV-FWDIVYPGDSFKIKTQLFTRTQPLNTAA 59
Query 66 FARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDG-------SMPF 118
+ R+REY DFYFVP L+ T+L QM DNP A S VV D S +
Sbjct 60 YTRIREYLDFYFVPLRLINKNLPTALMQMQDNPVQATGLSSNKVVTTDIPWVPTNLSGTY 119
Query 119 TDLSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYYYAESTSNTFAK 178
L++++ NS S+ST + + G++ SAKL+ L YGN S N
Sbjct 120 GSLTALADVKNSFPSSSTGI---EDLLGFDAITQSAKLLMYLRYGNFLSSVVSDKNKSLG 176
Query 179 RPLHYNL-----------NVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGS 227
+L ++ + L AYQK YAD++R +QWE+ P +N D+ YS G+
Sbjct 177 LSGSLDLRNSETASTGYTSMHILPLAAYQKAYADFFRFTQWEKNQPYTYNFDW--YSGGN 234
Query 228 SGMMLSFNYSDFYENY----SMFDLRYCNWQKDLFHGVVPNQQYGDVASI---------- 273
++ S D + Y ++F LRY NW KD+F GV+P+ Q GDV+ +
Sbjct 235 --VLASLTTLDLAKKYYSDDNLFTLRYANWPKDMFMGVMPDSQLGDVSIVDASGSEGTFP 292
Query 274 ---------SMSVPVVAGSSAAlinsritssnstttlrFPTDPAIPDATPLLTHPSFSIL 324
++ ++A S +A +++ L T + SFSIL
Sbjct 293 VGLLDVNDGTLRAGLLARSGSAPAEKSSLEMQTSSALSANTTYGVYAQRAAGLASSFSIL 352
Query 325 ALRQAEFLQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINEVVNQ 384
LR AE +QK++E++Q ++D + Q+ H+ VS S+ C YLGG SS++D++EVVN
Sbjct 353 QLRMAEAVQKYREVSQFADQDARGQIMAHFGVSLSPVLSDKCVYLGGSSSNIDLSEVVNT 412
Query 385 NITGSNAADIAGKGTGVSNGVINFNSQ-GRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNA 443
NITG N A+IAGKG G G +F+ Q YG+++ IYH +PL+DY ++ N
Sbjct 413 NITGDNVAEIAGKGVGTGQG--SFSGQFDEYGIIIGIYHNVPLLDYVITGQPQNLLYTNT 470
Query 444 ADFAIPEFDRVGMQTVPLSYVSNGPLSVLPLSIP---NEIGYAPRYIDYKTDIDTSIGAF 500
AD PEFD +GMQT+ N + +GY PR+ D KT D +GAF
Sbjct 471 ADLPFPEFDSIGMQTIQFGRFVNSKSVSWTSGVDYRVQTMGYLPRFFDVKTRYDEVLGAF 530
Query 501 KTSLKNWVISYDNQSLANQFGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEAD 560
+++LKNWV D ++ SV + N NY FKVNP L+ +F V+ D
Sbjct 531 RSTLKNWVAPLDPSYVSKWLQSSVTSSGKLALN-----LNYGFFKVNPRVLDSIFNVKCD 585
Query 561 SSIDTDQFLCSTFFDVKVVRNLDTDGLPY 589
S+IDTDQFL + + D+K VRN D DG+PY
Sbjct 586 STIDTDQFLTALYMDIKAVRNFDYDGMPY 614
> Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1
Length=622
Score = 370 bits (949), Expect = 4e-121, Method: Compositional matrix adjust.
Identities = 246/639 (38%), Positives = 357/639 (56%), Gaps = 67/639 (10%)
Query 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQP 60
MA+ LK L+N + GFD+ SK FTAK GEL+PV +P + IDL FTRT+P
Sbjct 1 MAHFTGLKELQNHPHKAGFDVGSKNLFTAKVGELIPVYWDFAIPDCDYDIDLAYFTRTRP 60
Query 61 LNTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTD 120
+ TAA+ R+REY+DFY VP DLLW ++++ QM + L+ + + +P+
Sbjct 61 VQTAAYTRVREYFDFYAVPCDLLWKSFDSAVIQM-GQTAPVQSKTLLDPLTVGNDIPWCT 119
Query 121 LSSISR--YLNS----LASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLY-------- 166
L +S Y +S L S + + N FGYNR +KL+ L YGN
Sbjct 120 LLDLSNAVYFSSGSSPLGSTVSVPSGFGNIFGYNRGDVDSKLLFYLNYGNFVNPSLSNVG 179
Query 167 ---------YYAESTSNTFAKRPLHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFN 217
++ S ++++ L+ N V++F LLAYQKIY D++R SQWE P+ +N
Sbjct 180 SPSNRWWNTSFSSSKVPGYSQKYLNNNA-VNIFPLLAYQKIYQDFFRWSQWENADPTSYN 238
Query 218 VDYMPYST---GSSGMMLSFNYSD-FYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASI 273
VDY S GSSG+ S S+ +++ +MF LRYCNW KD+F G++PN Q+GDVA +
Sbjct 239 VDYYNGSGNLFGSSGLSGSVTASNSYWKRDNMFSLRYCNWNKDMFTGLLPNSQFGDVAVV 298
Query 274 SM------SVPVVAGSSAAlinsritssnstttlrFPTDP-AIPDATPLLTH-------- 318
++ ++PV G + + ++ + T P I +TP+
Sbjct 299 NLGDSGSGTIPV--GFLSDTEVFTQAFNATSMSTVSDTSPMGISGSTPVSARQSMVARIN 356
Query 319 ----PSFSILALRQAEFLQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISS 374
SFSILALRQAE LQKWKEITQS + +Y++Q++ H+ ++ S M Y+GG++
Sbjct 357 NADVASFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMSHMAQYIGGVAR 416
Query 375 SLDINEVVNQNIT--GSNAADIAGKGTGVSNGVINFNSQGRYGVVMCIYHCLPLIDYTTD 432
+LDI+EVVN N+ GS A I GKG G +G + +++ +Y ++MCIYH +PL+DY
Sbjct 417 NLDISEVVNNNLKDDGSEAV-IYGKGVGSGSGKMRYHTGSQYCIIMCIYHAMPLLDYAIT 475
Query 433 FVSPSVTRVNAADFAIPEFDRVGMQTVPLSYVSNGPLSVL--PLSIPNEIGYAPRYIDYK 490
P + + D IPEFD +GM+ VP + + N SVL ++ + +GY PRY +K
Sbjct 476 GQDPQLLCTSVEDLPIPEFDNIGMEAVPATTLFN---SVLFDGTAVNDFLGYNPRYWPWK 532
Query 491 TDIDTSIGAFKTSLKNWVISYDNQSLANQFGYSVETPESPVPNPANSSWNYTLFKVNPNS 550
+ ID GAF T+LK+WV D+ L N F S A+ SW + FKVNPN+
Sbjct 533 SKIDRVHGAFTTTLKDWVAPIDDDYLHNWF-------NSKDGKSASISWPF--FKVNPNT 583
Query 551 LNPLFAVEADSSIDTDQFLCSTFFDVKVVRNLDTDGLPY 589
L+ +FAV ADS +TDQ L + KVVR L DG+PY
Sbjct 584 LDSIFAVVADSIWETDQLLINCDVSCKVVRPLSQDGMPY 622
> Alpavirinae_Human_feces_B_039_Microviridae_AG096_putative.VP1
Length=579
Score = 288 bits (737), Expect = 3e-90, Method: Compositional matrix adjust.
Identities = 204/616 (33%), Positives = 312/616 (51%), Gaps = 64/616 (10%)
Query 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEV-LPGDTFKIDLKSFTRTQ 59
M++ L + T R+ FDLSSK+ FTAK GE+LP W++ +PG ++I FTRT
Sbjct 1 MSDFNPLDRAKIPTHRSSFDLSSKKLFTAKVGEILPCY-WQIAIPGTKYRISSDWFTRTV 59
Query 60 PLNTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFT 119
P+NTAA+ R++EYYDFY VP L+ + TQM D A SS N L S+P
Sbjct 60 PVNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYMTSAA-SSTANTSALT-SVPSV 117
Query 120 DLSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYYYAESTSNTFAKR 179
S S + + +N+ N + G S KL++ LGYG++ + K+
Sbjct 118 TQSLFSAFFQT--ANAGDQPNTRDDAGLPIVYGSCKLLDMLGYGSMIDSKNTGKAAITKK 175
Query 180 ------------PLHY--NLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYST 225
PL Y + V+ LAYQKIY D++ ++QWE+ +NVDY T
Sbjct 176 YLGVDALGDADNPLVYQSSQTVNALPFLAYQKIYYDFFSNNQWEKHKAYAYNVDYWS-GT 234
Query 226 GSSGMMLSFNYSDFYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVASI-SMSVPVVAGSS 284
G+ G++ M LRY N+ KD F G++P+ QYG VA + S++ V+ +
Sbjct 235 GNIGLVTD-----------MVQLRYANYPKDYFMGMLPSSQYGSVAVLPSLTSAAVSPTF 283
Query 285 AAlinsritssnstttlrFPTDPAIPDATPLLTHPSF----SILALRQAEFLQKWKEITQ 340
N+ +S + + T L + S L+LR E+LQ+WKE+ Q
Sbjct 284 IINKNNVSSSYTIVNNNNNTLVASSTNTTGLEQYSQLNTDLSALSLRATEYLQRWKEVVQ 343
Query 341 SGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINEVVNQNI-TGSNAADIAGKGT 399
+KDY +Q+ + + + Y+GG SS ++INEVVN N+ T S+ A IAGKG
Sbjct 344 FSSKDYSDQMAAQFGIKAPEYMGNHAHYIGGWSSVININEVVNTNLDTDSSQASIAGKGV 403
Query 400 GVSNG-VINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSVTRVNAADFAIPEFDRVGMQT 458
++G I ++ + V+MC+YH +P++D+ +P +T +DF P FD++GMQ
Sbjct 404 SSNSGHTITYDCGAEHQVIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQA 463
Query 459 VPLSYVSNGPLSVLPLSIPNEIGYAPRYIDYKTDIDTSIGAFK--TSLKNWVISYDNQSL 516
VP + N P ++ +GY RY +K++IDT F+ + ++W D
Sbjct 464 VPALNLQNNP----GRNVSGSLGYNLRYWQWKSNIDTVHAGFRAGAAYQSWAAPLD---- 515
Query 517 ANQFGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLFAVEADS---SIDTDQFLCSTF 573
G+ V T A+ +W+Y K+ P LN +F + D+ S+ DQ LC+
Sbjct 516 ----GWQVLT--------ASGTWSYQSMKIRPQQLNSIFVPQIDAANCSVAFDQLLCNVN 563
Query 574 FDVKVVRNLDTDGLPY 589
F V V+NLD +GLPY
Sbjct 564 FQVYAVQNLDRNGLPY 579
> Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1
Length=618
Score = 198 bits (503), Expect = 7e-57, Method: Compositional matrix adjust.
Identities = 114/277 (41%), Positives = 163/277 (59%), Gaps = 22/277 (8%)
Query 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKAGELLPVKTWEVLPGDTFKIDLKSFTRTQP 60
M+++ S ++N R+GFDLS+K FTAK GELLPV LPGD F I + FTRTQP
Sbjct 1 MSSLFSYGDIKNSPRRSGFDLSNKCAFTAKVGELLPVYWKFCLPGDKFNISQEWFTRTQP 60
Query 61 LNTAAFARMREYYDFYFVPYDLLWNKANTSLTQMYDNPQHALDSSPLNVVKLDGSMPFTD 120
++T+AF R+REYY+++FVP LL+ +N ++ M + P +A S + + + ++P+ D
Sbjct 61 VDTSAFTRIREYYEWFFVPLHLLYRNSNEAIMSMENQPNYAASGS--SSISFNRNLPWVD 118
Query 121 LSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYGNLYYYAESTSNTFAKRP 180
LS+I N N + T+ N+FG +R+ KL+ LGYG E++ +
Sbjct 119 LSTI----NVAIGNVQSSTSPKNFFGVSRSEGFKKLVSYLGYG------ETSPEKYVD-- 166
Query 181 LHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGSSGMMLSFNYSDFY 240
NL S F L AYQKIY DYYR SQWE+ P +N D+ G ++ F
Sbjct 167 ---NLRCSAFPLYAYQKIYQDYYRHSQWEKSKPWTYNCDFW---NGEDSTPITATVELFS 220
Query 241 E--NYSMFDLRYCNWQKDLFHGVVPNQQYGDVASISM 275
+ N S+F+LRY NW KDL+ G +PN Q+GDVA IS+
Sbjct 221 QSPNDSVFELRYANWNKDLWMGSLPNSQFGDVAGISL 257
Score = 192 bits (487), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 113/274 (41%), Positives = 156/274 (57%), Gaps = 17/274 (6%)
Query 321 FSILALRQAEFLQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINE 380
FS+L LR AE LQKWKEI Q+ ++Y QV+ H+ VS S T + G S+DI+
Sbjct 357 FSVLQLRAAEALQKWKEIAQANGQNYAAQVKAHFGVSTNPMQSHRSTRICGFDGSIDISA 416
Query 381 VVNQNITGSNAADIAGKGTGVS--NGVINFNSQGRYGVVMCIYHCLPLIDYTTDFVSPSV 438
V N N+T ++ A I GKG G N +F +GV+MCIYH PL+DY P +
Sbjct 417 VENTNLT-ADEAIIRGKGLGGQRINDPSDFTCN-EHGVIMCIYHATPLLDYVP--TGPDL 472
Query 439 ---TRVNAADFAIPEFDRVGMQTVPLSYVSNGPLSVLPLSIPNEIGYAPRYIDYKTDIDT 495
+ V + +PEFD +GM+++P+ + N ++ + + GY PRYI +KT ID
Sbjct 473 QLMSTVKGESWPVPEFDSLGMESLPMLSLVNSK-AIGDIVARSYAGYVPRYISWKTSIDV 531
Query 496 SIGAFKTSLKNWVISYDNQSLANQFGYSVETPESPVPNPANSSWNYTLFKVNPNSLNPLF 555
GAF +LK+W D+ + FG + SP+ +YT FKVNP+ LNP+F
Sbjct 532 VRGAFTDTLKSWTAPVDSDYMHVFFGEVIPQEGSPI-------LSYTWFKVNPSVLNPIF 584
Query 556 AVEADSSIDTDQFLCSTFFDVKVVRNLDTDGLPY 589
AV D S +TDQ LC+ FDVKV RNL DG+PY
Sbjct 585 AVSVDGSWNTDQLLCNCQFDVKVARNLSYDGMPY 618
Lambda K H a alpha
0.317 0.133 0.401 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 54420456