bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
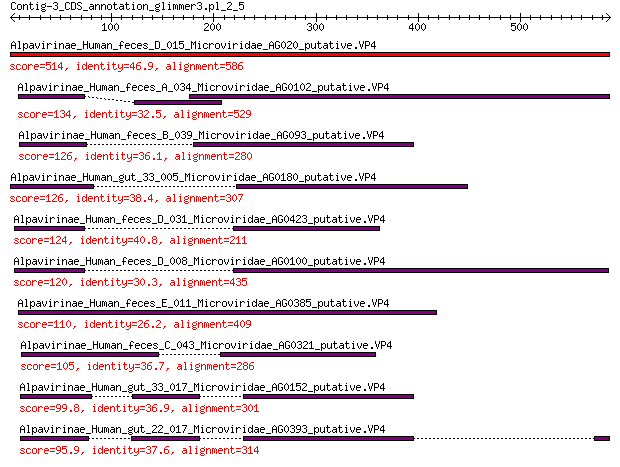
Query= Contig-3_CDS_annotation_glimmer3.pl_2_5
Length=587
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 514 8e-178
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 134 2e-35
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 126 8e-33
Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4 126 1e-32
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 124 5e-32
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 120 1e-30
Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4 110 3e-27
Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4 105 9e-26
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 99.8 8e-24
Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4 95.9 1e-22
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 514 bits (1323), Expect = 8e-178, Method: Compositional matrix adjust.
Identities = 275/586 (47%), Positives = 369/586 (63%), Gaps = 23/586 (4%)
Query 1 MADKVRNFFNRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCY 60
M KV +FNRCEHP++I+NKYTG+ V V+CG CP+CLI ++D CD+ K+N +YCY
Sbjct 1 MDKKVFKYFNRCEHPQVIQNKYTGDYVKVDCGQCPYCLIKKADRSTQKCDFVKYNHRYCY 60
Query 61 FVTLTYNSQYVPKMALVPITDYEFDYPIGKNWPCVKTQLHMRLMLDPRVKKTEMFSQETR 120
FVTLTYN+QYVPKM+L I DY ++ + TQL R++ D RV K
Sbjct 61 FVTLTYNTQYVPKMSLTQIEDYLSEWLPVRPPKSFGTQLTARMLTDSRVNK--------- 111
Query 121 MRARRPVGDSLINMFTCKVNKPYIDEHLKAIFDSCAAAVEFRKNYKPTYQSPARPYILRT 180
+ + + +VN+PY+ EHL+ + A+ R P + S ARPYILR+
Sbjct 112 ---------KIPDFMSAEVNRPYMLEHLRLLEADRYKALALRY---PNFISKARPYILRS 159
Query 181 IPRISKLQKFNDVQREELVWLSPEKVEMLKKKSKCEGDNNAFPQFKGLLKYINYRDYQLF 240
IPR+SKLQ F D EELVW+ PE E LKKK+ + N AFPQFKGLLKY+N RDYQLF
Sbjct 160 IPRVSKLQNFKDEYFEELVWMLPEIAESLKKKNNTDA-NGAFPQFKGLLKYVNIRDYQLF 218
Query 241 AKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEVAKNIRQAVFQSWKL 300
AKR RKYL ++G YEKI SYVVSEYSPKTFRPHFHILFFFDSDE+AKN RQAV+QSW+L
Sbjct 219 AKRLRKYLSKKVGKYEKIHSYVVSEYSPKTFRPHFHILFFFDSDEIAKNFRQAVYQSWRL 278
Query 301 GRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVSFTKNKSRFSKLFGYESFRQTIKTPEQ 360
GRVDTQLAR+ A SYVS YLNSVVS+P ++ + +SRFS LFG+E ++ I+ +
Sbjct 279 GRVDTQLAREQANSYVSNYLNSVVSIPFVYKAKKSIRPRSRFSNLFGFEEVKEGIRKAQD 338
Query 361 AIERLSERILFVRNGKPCEFTPPFSYISRLLPRFVPYSSNFVVETRTVLRSIRGVLQLFR 420
L + + ++ N K + P S I RL PRF Y +F+ + + ++ +L+LF
Sbjct 339 KRAALFDGLSYISNQKFVRYVPSGSLIDRLFPRFTYYDGSFLRRSSQIYGVVQQILRLFA 398
Query 421 RNEPFKKETPTNISEFVHTYTVTLYEKYGYCYDTLPECLRVYLSYTRSVKEIHYFTDKLK 480
RNEPF++ TP N+SEF+ + + + G P+ ++ +L R +E D K
Sbjct 399 RNEPFEEATPGNVSEFICWWCAYNFRQ-GCQIKDFPDYMQEFLHIVRLDRESFLNWDIPK 457
Query 481 NKLCRPLYIYRIWESLGLSDDCIISLSDEYMSRCRSMSLEKQLSIQQEMFEREGYSDELL 540
K+ R LY + ++E + S + + + SL+ QL +QQ +F GYSDELL
Sbjct 458 GKISRFLYRFNMFEKMKGSFRSKLKAVELFYDYRDYQSLKNQLYMQQLVFSELGYSDELL 517
Query 541 SLFYINKPQKKVNNRYFQEWKDKNYYEVHYIRVKHKKLNDENNVFL 586
FY+ K + N Y ++W D NY+EVHY RVKHK LND+N+VFL
Sbjct 518 DSFYVKPNLKVLKNIYTEKWHDTNYHEVHYFRVKHKVLNDQNDVFL 563
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 134 bits (336), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 130/447 (29%), Positives = 201/447 (45%), Gaps = 46/447 (10%)
Query 176 YILRTIPRISKLQKFNDVQREELVWLSPEKVEMLKKKSKCE----GDNNAF---PQFKGL 228
Y +T+ ++ FND + LS ++L+KK+ D P +GL
Sbjct 95 YSYKTVQGYTRNISFNDKTFDFDTQLSWSSAQLLQKKAHLHYTSFPDGRCIYNRPYMEGL 154
Query 229 LKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEVAK 288
+ Y+NY D QLF KR + + R + EKI YVV EY P TFRPHFHIL F DS ++ +
Sbjct 155 VGYLNYHDIQLFFKRLNQNI--RRITNEKIYYYVVGEYGPTTFRPHFHILLFHDSRKLRE 212
Query 289 NIRQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVSFTKNKSRFSKLFGY 348
+IRQ V +SW+ G DTQ SA YV+GY+NS LP F + K RFS F
Sbjct 213 SIRQFVSKSWRFGDSDTQPVWSSASCYVAGYVNSTACLPDFFKNSRHIKPFGRFSVNFAE 272
Query 349 ESFRQTIKTPEQAIERLS----ERILFVRNGKPCEFTPPFSYISRLLPRFVPYSSNFVVE 404
+F + K PE+ E S R+L + NGKP P S+I+RL PR VV+
Sbjct 273 SAFNEVFK-PEENEEIFSLFYDGRVLNL-NGKPTIVRPKRSHINRLYPRLDKSKHATVVD 330
Query 405 TRTVLRSIRGVLQLFRRNEPFKKETPTNISEFVHTYTVTLYEKYGYCYDTLPECLRVYLS 464
+ + + Q+ + + + +S+ V+ Y + + + D + LRV +
Sbjct 331 DIRIASVVSRLPQVVAKFGFIDEVSDFELSKRVY-YLIRRHLEVDCSLDYASDELRVIYN 389
Query 465 YTRSVKEIHY---------------FTDKLKNKLCRPLYIYRIWESLGLSDDCIISLSDE 509
R I++ + + +KN + P+ L + I S D
Sbjct 390 ACRLSLYINFSDESGCAAIYRLLLQYRNLVKNWITAPVGSVAFTGQLHRAVRAIHSFYD- 448
Query 510 YMSRCRSMSLEKQLSIQQEMFEREGYSDELLSLFY------INKPQKKVNNRYFQEWKDK 563
C SL QL ++ E + + Y LS++Y ++ K + F +
Sbjct 449 ---YCSRRSLHDQL-VKVEKWSNDSYVRANLSIYYFYPLTDVDIMMKSFSEIVFDSQVLR 504
Query 564 NYYEVHYI----RVKHKKLNDENNVFL 586
Y + R+KHK LND+N++ +
Sbjct 505 ASYADYAADNRERIKHKALNDKNSLLI 531
Score = 73.9 bits (180), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 28/64 (44%), Positives = 46/64 (72%), Gaps = 0/64 (0%)
Query 9 FNRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNS 68
+ C HPR+IKNKYTG+P+YV CG+C C+ +++ C+ + + +YC F+TLTY++
Sbjct 9 YGSCLHPRVIKNKYTGDPIYVPCGTCEFCVHNKAIKAELKCNVQLASSRYCEFITLTYST 68
Query 69 QYVP 72
+Y+P
Sbjct 69 EYLP 72
Score = 27.7 bits (60), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 36/88 (41%), Gaps = 3/88 (3%)
Query 122 RARRPVGDSLINMFTCKVNKPYIDEHLKAIFDSCAAAVEFRKNYKPTYQSPARPYILRTI 181
R +P G +N N+ + E + IF N KPT P R +I R
Sbjct 258 RHIKPFGRFSVNFAESAFNEVFKPEENEEIFSLFYDGRVLNLNGKPTIVRPKRSHINRLY 317
Query 182 PRISKLQK---FNDVQREELVWLSPEKV 206
PR+ K + +D++ +V P+ V
Sbjct 318 PRLDKSKHATVVDDIRIASVVSRLPQVV 345
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 126 bits (317), Expect = 8e-33, Method: Compositional matrix adjust.
Identities = 78/222 (35%), Positives = 119/222 (54%), Gaps = 7/222 (3%)
Query 180 TIPRISKLQKFNDVQREELVWLSPEKVEMLKKKSKCEGDNNAFPQF--KGLLKYINYRDY 237
T+P +++++ V+ + ++ + + K +S + D A Q+ L+ ++NY D
Sbjct 129 TVPFDREIKEYVPVKDNWFLSMAAIRSFIYKTQSVDKTDYPASEQYGRDNLIPFLNYVDV 188
Query 238 QLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEVAKNIRQAVFQS 297
Q + KR RKYLF ++G YE Y V EY P FRPH+H+L F +SD+V++ +R +S
Sbjct 189 QNYIKRLRKYLFQQLGKYETFHFYAVGEYGPVHFRPHYHLLLFTNSDKVSEVLRYCHDKS 248
Query 298 WKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVSFTKNKSR-----FSKLFGYESFR 352
WKLGR D Q + AGSYV+ Y+NS+ S P ++ + KSR F K +
Sbjct 249 WKLGRSDFQRSAGGAGSYVASYVNSLCSAPLLYRSCRAFRPKSRASVGFFEKGCDFVEDD 308
Query 353 QTIKTPEQAIERLSERILFVRNGKPCEFTPPFSYISRLLPRF 394
EQ I+ + ++ NG TPP SYI LLPRF
Sbjct 309 DPYAQIEQKIDSVVNGRVYNFNGVSVRSTPPLSYIRTLLPRF 350
Score = 54.7 bits (130), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 10 NRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNSQ 69
+ C+H I N+Y G + V+CG C +C+ R+ KY YFVTLTY+++
Sbjct 12 DHCQHRSFITNRYNGARITVDCGQCDYCIHKRAQKASMRVKTAGSAFKYSYFVTLTYDNE 71
Query 70 YVPKM 74
++P M
Sbjct 72 HIPLM 76
> Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4
Length=536
Score = 126 bits (316), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 85/230 (37%), Positives = 114/230 (50%), Gaps = 22/230 (10%)
Query 222 FPQFKGLLKYINYRDYQLFAKRFRKYLFTRIGSY--EKISSYVVSEYSPKTFRPHFHILF 279
+P LL Y+NYRD QLF KR + +I Y EKI SY V EY PKTFRPHFH+LF
Sbjct 147 YPNMGDLLPYLNYRDVQLFHKRINQ----QIKKYTDEKIYSYTVGEYGPKTFRPHFHLLF 202
Query 280 FFDSDEVAKNIRQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVSFTKNK 339
FFDS+ +A+ Q V ++W+ G DTQ SA SYV+GYLNS LP +
Sbjct 203 FFDSERLAQVFGQLVDKAWRFGNSDTQRVWSSASSYVAGYLNSSHCLPEFYRCNRKIAPF 262
Query 340 SRFSKLFGYESFRQTIKTP--EQAIERLSERILFVRNGKPCEFTPPFSYISRLLPRFVPY 397
RFS+ F F + K E+ ++ I GKP P S I+RL P
Sbjct 263 GRFSQYFAERPFIEAFKPEENEKVFDKFVNGIYLSLGGKPTLCRPKRSLINRLYP----- 317
Query 398 SSNFVVETRTVLRSIRGVLQLFRRNEPFKKETPTNISEFVHTYTVTLYEK 447
V R+ + + + R F + P +++F VTL+E+
Sbjct 318 -----VLDRSAVSDVDSNV----RTALFVSKIPQVLAKFGFLDEVTLFEQ 358
Score = 73.9 bits (180), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 33/81 (41%), Positives = 51/81 (63%), Gaps = 0/81 (0%)
Query 1 MADKVRNFFNRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCY 60
M + V + C HP IIKNKYTG+P+YVECG+C CL +++ K C+ + + + C+
Sbjct 1 MINPVVKPYCSCLHPVIIKNKYTGDPIYVECGTCEVCLSNKAIQKELRCNIQLASSRCCF 60
Query 61 FVTLTYNSQYVPKMALVPITD 81
FVTLTY ++++P + D
Sbjct 61 FVTLTYATEHIPVARFYKLND 81
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 124 bits (310), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 60/143 (42%), Positives = 85/143 (59%), Gaps = 1/143 (1%)
Query 219 NNAFPQFKGLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHIL 278
N +P G + Y+ + D L+ KR RKY+ +++G E I +Y+V EY P +FRPHFH+L
Sbjct 147 NGKYPALSGRIPYLLHGDVSLYMKRVRKYI-SKLGINETIHTYIVGEYGPSSFRPHFHLL 205
Query 279 FFFDSDEVAKNIRQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVSFTKN 338
FFDSDE+A+NI + W+ GRVD +R A YVS YLNS S+P ++ +
Sbjct 206 LFFDSDELAQNIIRIASSCWRFGRVDCSASRGDAEDYVSSYLNSFSSIPLHIQEIRAIRP 265
Query 339 KSRFSKLFGYESFRQTIKTPEQA 361
+RFS FGY F +IK +
Sbjct 266 FARFSNKFGYSFFESSIKKAQSG 288
Score = 60.8 bits (146), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 40/68 (59%), Gaps = 0/68 (0%)
Query 5 VRNFFNRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTL 64
++ F+ C+H KN YTGE + V CG+CP C ++S +N + ++ YFVTL
Sbjct 7 IQKVFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLVSRHVYFVTL 66
Query 65 TYNSQYVP 72
TY +Y+P
Sbjct 67 TYAQRYIP 74
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 120 bits (301), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 106/384 (28%), Positives = 182/384 (47%), Gaps = 26/384 (7%)
Query 219 NNAFPQFKGLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHIL 278
+ +P + Y+ + D L+ KR RKY+ +++G E I +YVV EY P TFRPHFH+L
Sbjct 147 DGKYPALSDRIPYLLHDDVSLYMKRVRKYI-SKLGINETIHTYVVGEYGPVTFRPHFHLL 205
Query 279 FFFDSDEVAKNIRQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVSFTKN 338
FF+SDE+A++I + W+ GRVD +R A YVS YLNS S+P ++ +
Sbjct 206 LFFNSDELAQSIVRIARSCWRFGRVDCSASRGDAEDYVSSYLNSFSSIPLHIQEIRAIRP 265
Query 339 KSRFSKLFGYESFRQTIKTPEQA-IERLSERILFVRNGKPCEFTPPFSYISRLLPRFVPY 397
+RFS FG+ F +IK + + + NG P + I R
Sbjct 266 FARFSNKFGFSFFESSIKKAQSGDFDEILNGKSLPYNGFNTTIFPWRAIIDTCFYRPALR 325
Query 398 SSNFVVETRTVLRSIRGVLQLFRRNEPFKKETPTNISEFVHTYTVTL-------YEKYGY 450
+ + E +LR +R F++ F+K T ++TY L + + Y
Sbjct 326 RRSDIHELTEILRYVRN----FKQRPAFQKATLFQSPRIMYTYLQDLGPSAAAKFVESDY 381
Query 451 CYDTLPECLRVYLSYTRSVK----EIHYFTDKLKNKLCRP-LYIYRIWESLGLSDDCIIS 505
+ L+ L YT+ ++ E+ F +L L + L++ I +L LS S
Sbjct 382 PLHRILSFLK--LDYTKIIQGEPSEVQSFHSRLYMLLRQSELFLNGIGYTL-LSTRVEYS 438
Query 506 LSDEYMSRCRSMSLEKQLSIQQEMF-EREGYSDELLSLFYINKPQKK---VNNRYFQEWK 561
L + + + E++ Q++F + E + + +F+ + +K V++ Y +
Sbjct 439 LIKKSLENSINFYNERERKSLQDLFHDSEAFESDWSDIFWDRRQEKIKRFVDSDYGNLCR 498
Query 562 DKNYYEVHYIRVKHKKLNDENNVF 585
DK + E+ R+KH+++ND +F
Sbjct 499 DKLHNEIRK-RIKHREINDAVGIF 521
Score = 60.8 bits (146), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 40/68 (59%), Gaps = 0/68 (0%)
Query 5 VRNFFNRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTL 64
++ F+ C+H KN YTGE + V CG+CP C ++S +N + ++ YFVTL
Sbjct 7 IQKVFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLISRHVYFVTL 66
Query 65 TYNSQYVP 72
TY +Y+P
Sbjct 67 TYAQRYIP 74
> Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4
Length=611
Score = 110 bits (275), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 107/418 (26%), Positives = 188/418 (45%), Gaps = 50/418 (12%)
Query 9 FNRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNS 68
F C P+ I N Y+ + ++ CG C C++++S+ + KYCYFVTLTY
Sbjct 13 FTECLRPQRIVNPYSHDVIFAPCGHCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYKD 72
Query 69 QYVPKMALVPITDYEFDYPIGKNWPCVKTQLHMRLMLDPRVKKTEMFSQETRMRARRPVG 128
++P +++ + Y +N+ M DPR E +
Sbjct 73 IFLPYLSVEVVRRSGNRYLFDENFET------MVSTSDPRFLTPEYYHDRD--------- 117
Query 129 DSLINMFTCKVNKPYIDEHLKAIFDSCAAAVEFRKNYKPTYQSPARPYILRTIP-RISKL 187
F+ + +++ F S V + ++S + IP ++++L
Sbjct 118 ------FSLDSAQNEVEQVFDVGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTEL 171
Query 188 QKFNDVQREELVWLSPEKVEMLKKKSKCEGDNNAFPQF---KGLLKYINYRDYQLFAKRF 244
Q +++ + + + KKK +P K + + RD +LF KR
Sbjct 172 Q--------DILIKANGRYDYGKKKV-------VYPSLADCKLQIPVLQSRDIELFFKRL 216
Query 245 RKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEVAKNIRQAVFQSWKLGRVD 304
R+ L + + KI YVVSEY P+T+RPH+H L FF+S+E+ + +R+ + ++W GR+D
Sbjct 217 RRNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFFNSEEITQTLREDISKAWSYGRID 276
Query 305 TQLARDSAGSYVSGYLNSVVSLPGIFTDVSFTKNKSRFSKLFGYES-FRQTIKTPEQAIE 363
L+R +A SYV+ Y+NS LP ++ + +S SK FG F ++ E I
Sbjct 277 YSLSRGAAASYVAAYVNSAACLPFLYVGQKEIRPRSFHSKGFGSNKVFPKSSDVSE--IS 334
Query 364 RLSERIL----FVRNGKPCEFTPPFSYISRLLPRFVPYSSNFVVETRTVLRSIRGVLQ 417
++S+ NGK P S + PRF S++F ++ T + + V++
Sbjct 335 KISDLFFDGVNVDSNGKVVNIRPVRSCELAVFPRF---SNDFFSDSDTCCKLFQSVIE 389
> Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4
Length=546
Score = 105 bits (262), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 61/152 (40%), Positives = 84/152 (55%), Gaps = 3/152 (2%)
Query 206 VEMLKKKSKCEGDNNAFPQFKGLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSE 265
+E KK++ + +P K + Y++ +D QLF KR RK + R + EKI +Y+V E
Sbjct 130 IENFKKQANLDV-KGCYPHLKDMYGYLSRKDCQLFMKRVRKQI--RNYTDEKIHTYIVGE 186
Query 266 YSPKTFRPHFHILFFFDSDEVAKNIRQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVS 325
YSPK FRPHFHILFFF+S+E++++ V WK G VD +R A SYV+GY+NS
Sbjct 187 YSPKHFRPHFHILFFFNSNELSQSFGSIVRSCWKFGLVDWSQSRGDAESYVAGYVNSFAR 246
Query 326 LPGIFTDVSFTKNKSRFSKLFGYESFRQTIKT 357
LP RFS F F +T
Sbjct 247 LPYHLGSSPKIAPFGRFSNHFSESCFDDAKQT 278
Score = 54.7 bits (130), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 44/136 (32%), Positives = 63/136 (46%), Gaps = 21/136 (15%)
Query 12 CEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNSQYV 71
C H + + NKYTG+ +Y CG C CL + A+ + KY FVTLTY++ +V
Sbjct 15 CLHGKNVYNKYTGQMMYQSCGKCEACLSRLASARSIKVGVQASLSKYVMFVTLTYDTYHV 74
Query 72 PKMALVPITDYEFDYPIGKNWPCVKTQLHMRLMLDPRVKKTEMFSQETRMRARRPVGDSL 131
PK + Y G N + L++ PRVK + F ET +R G S
Sbjct 75 PKCKI---------YSNGDN--------NYVLVVKPRVK--DYFYYETSDGQKRLKGLSY 115
Query 132 INMFTC--KVNKPYID 145
+ F K +K YI+
Sbjct 116 DDDFRVEFKADKDYIE 131
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 99.8 bits (247), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 60/170 (35%), Positives = 91/170 (54%), Gaps = 9/170 (5%)
Query 229 LKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEVAK 288
+ YI RD LF KR R Y YEK+ Y VSEY P ++RPH+H+L F +S++ +K
Sbjct 163 IPYICNRDLDLFLKRLRSYY-----PYEKLRYYAVSEYGPTSYRPHWHLLLFSNSEQFSK 217
Query 289 NIRQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVS-FTKNKSRFSKLFG 347
I + V ++W GR D L+R A YV+ Y+NS V+LP +T++ F + KS S F
Sbjct 218 TILENVSKAWSYGRCDASLSRGFAAPYVASYVNSFVALPSFYTEMPRFLRPKSFHSIGFT 277
Query 348 YESF---RQTIKTPEQAIERLSERILFVRNGKPCEFTPPFSYISRLLPRF 394
+ + I ++ ++ + R+G P + Y+ RL PRF
Sbjct 278 ESNLFPRKVRISEIDEVTDKCLNGVRVERDGYFRILKPSWPYLLRLFPRF 327
Score = 69.3 bits (168), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 31/69 (45%), Positives = 43/69 (62%), Gaps = 0/69 (0%)
Query 11 RCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNSQY 70
RC++PR + NKYT EPV V CG+CP C++ RS + NL +Y YFVTLTY +
Sbjct 13 RCQNPRTVVNKYTHEPVVVSCGACPSCVLRRSGIQTNLLTTYSAQFRYVYFVTLTYAPCF 72
Query 71 VPKMALVPI 79
+P + + I
Sbjct 73 LPTLEVSVI 81
Score = 28.9 bits (63), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 30/66 (45%), Gaps = 4/66 (6%)
Query 120 RMRARRPVGDSLINMFTCKVNKPYIDEHLKAIFDSCAAAVEFRKNYKPTYQSPARPYILR 179
R ++ +G + N+F KV IDE + D C V ++ P+ PY+LR
Sbjct 267 RPKSFHSIGFTESNLFPRKVRISEIDE----VTDKCLNGVRVERDGYFRILKPSWPYLLR 322
Query 180 TIPRIS 185
PR S
Sbjct 323 LFPRFS 328
> Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4
Length=562
Score = 95.9 bits (237), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 58/172 (34%), Positives = 90/172 (52%), Gaps = 13/172 (8%)
Query 229 LKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEVAK 288
+ YI RD LF KR R Y EK+ Y VSEY P +FRPH+H+L F +S+ ++
Sbjct 163 IPYICNRDLDLFLKRLRSYYLD-----EKLRYYAVSEYGPTSFRPHWHLLLFSNSERFSR 217
Query 289 NIRQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVSFTKNKSRFSKLFGY 348
+ + V ++W GR D L+R A YV+ Y+NS V+LP +T + F +
Sbjct 218 TVCENVSKAWSYGRCDASLSRGFAAPYVASYVNSFVALPDFYTQMPKVVRPKSFHSIGFT 277
Query 349 ES--FRQTIKTPEQAIERLSERIL----FVRNGKPCEFTPPFSYISRLLPRF 394
ES F + ++ E ++ ++++ L R+G P + Y+ RL PRF
Sbjct 278 ESNLFPRKVRVAE--VDEITDKCLNGVRVERDGYFRTIKPTWPYLLRLFPRF 327
Score = 73.2 bits (178), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 31/66 (47%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 11 RCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNSQY 70
RC+HPR + NKYT EPV V CG CP C++ RS + NL +Y YFVTLTY +
Sbjct 13 RCQHPRTVVNKYTHEPVVVSCGHCPSCILRRSSVQTNLLTTYSAQFRYVYFVTLTYAPSF 72
Query 71 VPKMAL 76
+P + +
Sbjct 73 LPTLEV 78
Score = 28.5 bits (62), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 20/67 (30%), Positives = 29/67 (43%), Gaps = 4/67 (6%)
Query 119 TRMRARRPVGDSLINMFTCKVNKPYIDEHLKAIFDSCAAAVEFRKNYKPTYQSPARPYIL 178
R ++ +G + N+F KV +DE I D C V ++ P PY+L
Sbjct 266 VRPKSFHSIGFTESNLFPRKVRVAEVDE----ITDKCLNGVRVERDGYFRTIKPTWPYLL 321
Query 179 RTIPRIS 185
R PR S
Sbjct 322 RLFPRFS 328
Score = 24.6 bits (52), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 9/15 (60%), Positives = 13/15 (87%), Gaps = 0/15 (0%)
Query 572 RVKHKKLNDENNVFL 586
+VKHKK+ND + +FL
Sbjct 548 KVKHKKVNDLSGIFL 562
Lambda K H a alpha
0.323 0.138 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 54209524