bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_2
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
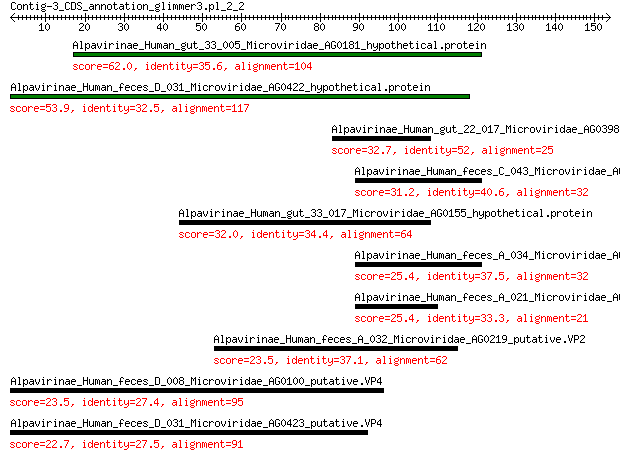
Alpavirinae_Human_gut_33_005_Microviridae_AG0181_hypothetical.p... 62.0 2e-14
Alpavirinae_Human_feces_D_031_Microviridae_AG0422_hypothetical.... 53.9 9e-12
Alpavirinae_Human_gut_22_017_Microviridae_AG0398_hypothetical.p... 32.7 2e-04
Alpavirinae_Human_feces_C_043_Microviridae_AG0323_hypothetical.... 31.2 6e-04
Alpavirinae_Human_gut_33_017_Microviridae_AG0155_hypothetical.p... 32.0 0.001
Alpavirinae_Human_feces_A_034_Microviridae_AG0103_hypothetical.... 25.4 0.055
Alpavirinae_Human_feces_A_021_Microviridae_AG078_hypothetical.p... 25.4 0.066
Alpavirinae_Human_feces_A_032_Microviridae_AG0219_putative.VP2 23.5 0.83
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 23.5 1.00
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 22.7 1.8
> Alpavirinae_Human_gut_33_005_Microviridae_AG0181_hypothetical.protein
Length=205
Score = 62.0 bits (149), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 37/104 (36%), Positives = 54/104 (52%), Gaps = 5/104 (5%)
Query 17 VLHTKVPVQDKLMQLSTFVNKDGSIAISSDISLIFNQQRLENKLTASELREYIQRYTPNK 76
+L PV D L N DGS+ SD ++F Q+ ++N + +LR Y+ P
Sbjct 79 ILPEPNPVGDFLFDH----NADGSVTFCSDYGILFGQKAIDN-MNQVQLRRYMNSLVPRS 133
Query 77 SVYTSQLDDDTLLNTLKSRHIQSLSEMRSWAEYCMENYDSLIKD 120
S YT +DD LL+ K R+IQS +EM SW ++ + SL D
Sbjct 134 SNYTRNYNDDFLLDYCKDRNIQSATEMASWLDHLLSEGQSLESD 177
> Alpavirinae_Human_feces_D_031_Microviridae_AG0422_hypothetical.protein
Length=150
Score = 53.9 bits (128), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 63/122 (52%), Gaps = 5/122 (4%)
Query 1 MKKKEYIDHIF-NGSFD---VLHTKVPVQDKLMQLSTFV-NKDGSIAISSDISLIFNQQR 55
M+K+E D I + D VL + + Q +++ +V + DG I SD++L+ N +R
Sbjct 1 MEKEEKKDKIIETPALDTARVLKSAIYCQVGPVEMLRYVKDDDGVIRYVSDVNLLMNAER 60
Query 56 LENKLTASELREYIQRYTPNKSVYTSQLDDDTLLNTLKSRHIQSLSEMRSWAEYCMENYD 115
L N++ I+ P KS Y ++ D+ L +KSR IQ+ SE+ +W E D
Sbjct 61 LRNQIGEESYLNLIRGIQPKKSPYDNKYTDEQLFTAIKSRFIQTPSEVLAWIESLGSAGD 120
Query 116 SL 117
S+
Sbjct 121 SI 122
> Alpavirinae_Human_gut_22_017_Microviridae_AG0398_hypothetical.protein
Length=69
Score = 32.7 bits (73), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 13/25 (52%), Positives = 20/25 (80%), Gaps = 0/25 (0%)
Query 83 LDDDTLLNTLKSRHIQSLSEMRSWA 107
+ DD LL T++SRHIQ+ SE+ +W+
Sbjct 4 MSDDDLLATVRSRHIQAPSEIIAWS 28
> Alpavirinae_Human_feces_C_043_Microviridae_AG0323_hypothetical.protein
Length=59
Score = 31.2 bits (69), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 13/32 (41%), Positives = 22/32 (69%), Gaps = 0/32 (0%)
Query 89 LNTLKSRHIQSLSEMRSWAEYCMENYDSLIKD 120
+ T+KSR++QS SE+R+W E ++ D + D
Sbjct 1 METIKSRYLQSPSEVRAWLETLVDKADVVRSD 32
> Alpavirinae_Human_gut_33_017_Microviridae_AG0155_hypothetical.protein
Length=171
Score = 32.0 bits (71), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/67 (33%), Positives = 40/67 (60%), Gaps = 4/67 (6%)
Query 44 SSDISLIFNQQRLENKL---TASELREYIQRYTPNKSVYTSQLDDDTLLNTLKSRHIQSL 100
+SDI LI + + L ++ AS+ + Q + + + + D+D LL T++SR+IQS
Sbjct 65 TSDIRLILHNKDLASRAGVDVASKFGQSKQSPSQIQQIMDTMSDED-LLATVRSRYIQSP 123
Query 101 SEMRSWA 107
SE+ +W+
Sbjct 124 SEILAWS 130
> Alpavirinae_Human_feces_A_034_Microviridae_AG0103_hypothetical.protein
Length=62
Score = 25.4 bits (54), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 12/32 (38%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 89 LNTLKSRHIQSLSEMRSWAEYCMENYDSLIKD 120
+ K R+IQS +EM++W E+ + SL D
Sbjct 1 MEYCKDRNIQSYTEMQAWLEHLISEGQSLEGD 32
> Alpavirinae_Human_feces_A_021_Microviridae_AG078_hypothetical.protein
Length=63
Score = 25.4 bits (54), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 7/21 (33%), Positives = 18/21 (86%), Gaps = 0/21 (0%)
Query 89 LNTLKSRHIQSLSEMRSWAEY 109
++ +K R++QS +E+++W+E+
Sbjct 1 MSLIKPRNVQSHAELKAWSEF 21
> Alpavirinae_Human_feces_A_032_Microviridae_AG0219_putative.VP2
Length=355
Score = 23.5 bits (49), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 23/68 (34%), Positives = 31/68 (46%), Gaps = 7/68 (10%)
Query 53 QQRLEN-KLTASELRE---YIQRYTPNKSVYT--SQLDDDTLLNTLKSRHIQSLSEMRSW 106
+Q++EN K TA E IQ NK V T +Q DD + N +KS + W
Sbjct 191 EQKIENLKKTAQATEESIKLIQANVANKEVQTRIAQFQDD-MNNIIKSSVWYEDGKSHGW 249
Query 107 AEYCMENY 114
E +E Y
Sbjct 250 EETVVERY 257
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 23.5 bits (49), Expect = 1.00, Method: Compositional matrix adjust.
Identities = 26/105 (25%), Positives = 40/105 (38%), Gaps = 20/105 (19%)
Query 1 MKKKEYIDHIFNGSFDVLHTKVPVQDKLMQLSTFVNKDGSIAISSDISLIFNQQRL-ENK 59
M ++E+I +F+ + TK P +L+ + + + FN+ L +NK
Sbjct 1 MNQQEFIQKVFSMCQHQVQTKNPYTGELIM----------VPCGTCPACRFNKSILSQNK 50
Query 60 LTASEL---------REYIQRYTPNKSVYTSQLDDDTLLNTLKSR 95
+ A L Y QRY P LD D L T R
Sbjct 51 VHAQSLISRHVYFVTLTYAQRYIPYYEYEIEALDADFLAITAHCR 95
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 22.7 bits (47), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 25/101 (25%), Positives = 39/101 (39%), Gaps = 20/101 (20%)
Query 1 MKKKEYIDHIFNGSFDVLHTKVPVQDKLMQLSTFVNKDGSIAISSDISLIFNQQRL-ENK 59
M ++E+I +F+ + TK P +L+ + + + FN+ L +NK
Sbjct 1 MNQQEFIQKVFSMCQHQVQTKNPYTGELIM----------VPCGTCPACRFNKSILSQNK 50
Query 60 LTASELRE---------YIQRYTPNKSVYTSQLDDDTLLNT 91
+ A L Y QRY P LD D L T
Sbjct 51 VHAQSLVSRHVYFVTLTYAQRYIPYYEYEIEALDADFLAIT 91
Lambda K H a alpha
0.316 0.130 0.361 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 10172981