bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_4
Length=365
Score E
Sequences producing significant alignments: (Bits) Value
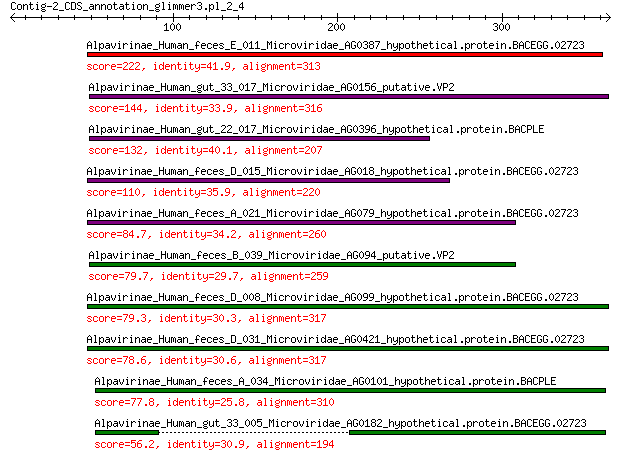
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 222 9e-70
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 144 4e-41
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 132 1e-36
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 110 1e-28
Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.p... 84.7 6e-20
Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2 79.7 3e-18
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 79.3 4e-18
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 78.6 7e-18
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 77.8 1e-17
Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.p... 56.2 1e-10
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 222 bits (566), Expect = 9e-70, Method: Compositional matrix adjust.
Identities = 131/316 (41%), Positives = 188/316 (59%), Gaps = 12/316 (4%)
Query 48 DFNAREAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYDS-NTGVagstgsts 106
+FNAREA+KAR +Q EMWN+ N++NSP N RK EAGYNPYLG DS N G A S GS+S
Sbjct 85 EFNAREAEKARQYQSEMWNKTNDWNSPKNVRKRLQEAGYNPYLGLDSSNVGTAQSAGSSS 144
Query 107 qaqaasPLSLNPEVYSELGSQLGRAGQMIYQERESNARTKALQGDADVARAQALQVFSNV 166
A AA P+ NP + + L A QM + SNA LQG +A AQA S +
Sbjct 145 PASAAPPIQNNPIQFDGFQNALSTAIQMSNSTKVSNAEANNLQGQKGLADAQAAATLSGI 204
Query 167 DWGKLSPDYKKWMRETGLQRAQLDYDTNKQNLQNLRWSNLIQMAERTNLLLSANTKRTLN 226
DW K +P+Y+ W++ TG+ RAQL ++T++QNL+N++W N IQ A+RT++LLS K +N
Sbjct 205 DWYKFTPEYRAWLQTTGMARAQLSFNTDQQNLENMKWVNKIQRAQRTDILLSNQAKGIIN 264
Query 227 KYLDQSEQTRINVMAAQYYDLMAAGHLKYQQCKESIAKQILYSKQGSWYDSMANKNNLDY 286
KYLD S+ ++ +MA Q + A+G L QQCK + KQ++ +MA
Sbjct 265 KYLDTSQSLQLKLMANQSFQAFASGRLSLQQCKTEVTKQLM---------NMAETEGKKI 315
Query 287 RNALA--LADDYIAAMSTQYESQTAYNMGFGQKAQEAGRRDAESKSFKSLIDRWNYNKRY 344
N +A AD I A+ QY + Y+ G+ A+EAG+ + K +D ++Y+ RY
Sbjct 316 SNKIASETADQLIGALQWQYSADEMYSRGYAGYAREAGKSRGKGDVAKGQLDEYDYSSRY 375
Query 345 YEEGLNTLQVVANAIG 360
+ G+ ++ + N IG
Sbjct 376 WNTGIESINRIGNGIG 391
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 144 bits (364), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 107/318 (34%), Positives = 168/318 (53%), Gaps = 16/318 (5%)
Query 49 FNAREAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYDS-NTGVagstgstsq 107
FN R A + RDFQ MWN++N YN+ + QR+ EAG NPYL + ++GV+ S G+ +
Sbjct 45 FNERMAMQQRDFQENMWNKENTYNTASAQRQRLEEAGLNPYLMMNGGSSGVSQSAGTGAS 104
Query 108 aqaasPLSLNPEVYSELGSQLGRAGQMIYQERESNARTKALQGDADVARAQALQVFSNVD 167
A ++ P + S + +A ++Q + A+ +QG ++A AQA+Q S VD
Sbjct 105 ASSSGTAVFQP--FQADFSGIQQAIGSVFQSQVRQAQVSQMQGQRNLADAQAMQALSQVD 162
Query 168 WGKLSPDYKKWMRETGLQRAQLDYDTNKQNLQNLRWSNLIQMAERTNLLLSANTKRTLNK 227
W K++ + +++++ TGL RA+L Y Q L N+ ++ + A+ T+ LL A+ K LN+
Sbjct 163 WSKMTKETREYLKATGLARARLGYSKEMQELDNMAFAGRLLQAQGTSQLLEADAKTVLNR 222
Query 228 YLDQSEQTRINVMAAQYYDLMAAGHLKYQQCKESIAKQIL-YSKQGSWYDSMANKNNLDY 286
YLDQ +Q +NV A+ YY+ M+ GHL Y Q K+ IA +IL Y++ L
Sbjct 223 YLDQQQQADLNVKASVYYNQMSQGHLNYNQAKKVIADEILTYAR--------IKGQKLSN 274
Query 287 RNALALADDYIAAMSTQYESQTAYNMGFGQKAQEAGRRDAESKSFKSLIDRWNYNKRYYE 346
+ A A AD I A + S N F +A + R A S+S + N K+Y
Sbjct 275 KVAEATADSLIRATNAANRS----NAEFDLEAAKYNRERARSRSIEDWYRSRNEGKKYKY 330
Query 347 EGLNTLQVVANAIGSIRG 364
+ L +IG+ G
Sbjct 331 YDSDKLIHYGTSIGNTVG 348
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 132 bits (333), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 83/210 (40%), Positives = 124/210 (59%), Gaps = 7/210 (3%)
Query 49 FNAREAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYDSNT-GVagstgstsq 107
FN R A + R++Q MWN++N YN+ + QR+ EAG NPYL + + GVA S G+ S
Sbjct 45 FNERMAIQQRNWQENMWNKENAYNTASAQRQRLEEAGLNPYLMMNGGSAGVAQSAGTGSA 104
Query 108 aqaasPLSLNP--EVYSELGSQLGRAGQMIYQERESNARTKALQGDADVARAQALQVFSN 165
A ++ + P YS +GS +G I+Q + LQG +A A+A++ SN
Sbjct 105 ASSSGNAVMQPFQADYSGIGSSIGN----IFQYELMQSEKSQLQGARQLADAKAMETLSN 160
Query 166 VDWGKLSPDYKKWMRETGLQRAQLDYDTNKQNLQNLRWSNLIQMAERTNLLLSANTKRTL 225
+DWGKL+ + + +++ TGL RAQL Y +Q N+ + L+ A+R+ +LL K L
Sbjct 161 IDWGKLTDETRGFLKSTGLARAQLGYAKEQQEADNMAMTGLVLRAQRSGMLLDNEAKGIL 220
Query 226 NKYLDQSEQTRINVMAAQYYDLMAAGHLKY 255
NKYLDQ +Q ++V AA YY MAAG++ Y
Sbjct 221 NKYLDQHQQLDLSVKAADYYQRMAAGYVSY 250
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 110 bits (274), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 79/224 (35%), Positives = 125/224 (56%), Gaps = 9/224 (4%)
Query 48 DFNAREAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYDSNTGVagstgstsq 107
+FNA+EA+KAR FQL+MWN++N YN+PA QR E GYN Y+ ++ G A STS
Sbjct 71 EFNAKEAEKARAFQLDMWNKENAYNTPAAQRARLEEGGYNAYMN-PADAGSASGMSSTSA 129
Query 108 aqaasPLSLNPEVYSELGSQLGRAGQMIYQERESNARTKALQ----GDADVARAQALQVF 163
A AAS + +S LG G + QE ++ + K L D ++Q ++
Sbjct 130 ASAASSAVMQGTDFSSLGE----VGVRLAQELKTFSEKKGLDIRNFSLKDYLQSQIDKMK 185
Query 164 SNVDWGKLSPDYKKWMRETGLQRAQLDYDTNKQNLQNLRWSNLIQMAERTNLLLSANTKR 223
+ +W +SP+ ++ +GL+ A++ + ++ N WSN + A N LL A +K
Sbjct 186 GDTNWRNVSPEAIRYNIMSGLEAAKIGMENLREQWANQVWSNNLLRANVANSLLDAESKT 245
Query 224 TLNKYLDQSEQTRINVMAAQYYDLMAAGHLKYQQCKESIAKQIL 267
LNKYLDQ +Q +NV AA Y +L+ G L + +E +++++L
Sbjct 246 ILNKYLDQQQQADLNVKAAHYEELINRGQLHVVEARELLSREVL 289
> Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.protein.BACEGG.02723
Length=397
Score = 84.7 bits (208), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 89/293 (30%), Positives = 136/293 (46%), Gaps = 57/293 (19%)
Query 48 DFNAREAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYD-SNTGVagstgsts 106
+F + EAQK RDF+++MWN+ NEYNS NQR EAG NPY+ + N G AGS + S
Sbjct 18 NFQSAEAQKNRDFEVDMWNKTNEYNSATNQRARLEEAGLNPYMMMNGGNAGEAGSVTAPS 77
Query 107 qaqaasPLS---LNPEVYSELGSQLGRAGQ----MIYQERES------------------ 141
Q A P + V S L S GQ M+ Q R +
Sbjct 78 TPQGAMPGATGDTTENVISGLNSVSNAIGQFYDNMLTQSRATAQNYENFFNDPSSYGKDQ 137
Query 142 --NARTKALQGDAD----VARAQALQVFSNVDWGKLSPDYKKWMRETGLQRAQLDYDTNK 195
K L D+D +++ A ++F +G M + +Q + D D
Sbjct 138 WQAMMMKMLSPDSDNSPFLSKDSASRIFGK--YGNWKQAGSAVMFDNAVQSS--DLDRQN 193
Query 196 QNLQN-LRWSNLIQMAERTNLLLSANTKRTLNKYLDQSEQTRINVMAAQYYDLMAAGHLK 254
+NL N + +N+IQ+ LS++ ++ +NKYLDQ E ++N+ +A Y + A G L
Sbjct 194 KNLTNQMIQANMIQVN------LSSDAQKVINKYLDQQESVKLNIQSALYTEAAARGQLT 247
Query 255 YQQCKESIAKQILYSKQGSWYDSMANKNNLDYRNALALADDYIAAMSTQYESQ 307
++Q QG +M+++ LDY A+AD++I A Y+ Q
Sbjct 248 FEQW------------QGQLIQNMSDR--LDYNTRKAIADEFIRASCEAYKLQ 286
> Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2
Length=380
Score = 79.7 bits (195), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 77/271 (28%), Positives = 125/271 (46%), Gaps = 31/271 (11%)
Query 49 FNAREAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYDSNTGVagstgstsqa 108
FNA +AQ RD+Q +MW N YNSP++ G NP++ + + S S A
Sbjct 41 FNAEQAQIQRDWQKQMWGMNNAYNSPSSM----ISRGLNPFVQGSAAMAGSKSPASGGAA 96
Query 109 qaasPLSLNPEVYSELGSQLGRAGQMIYQERESNARTKALQGDADVARAQALQ------V 162
A+P+ S ++Q S A+ KA + A + ++A Q +
Sbjct 97 ATAAPVPSMQAYKPNFSS--------VFQSLASLAQAKASEASAGESGSRARQTDTVTPL 148
Query 163 FSN-----VDWGKLSPDYKK-WMRETGLQRAQLDYDTNKQNLQNLRWSNLIQMAERTNLL 216
S+ +W L+ W +ETG A LD T QNL+N +++ I A+ +L
Sbjct 149 LSDYYRGLTNWKNLAIGSSGYWNKETGRVSAALDQSTETQNLKNAQFAERISAAQEAQIL 208
Query 217 LSANTKRTLNKYLDQSEQTRINVMAAQYYDLMAAGHLKYQQCKESIAKQILYSKQGSWYD 276
L+++ +R +NKY+DQ++Q + + A +L + G L +Q + I + IL S +
Sbjct 209 LNSDAQRIMNKYMDQNQQADLFIKAQTLANLQSQGALTEKQIQTEIQRAILASAE----- 263
Query 277 SMANKNNLDYRNALALADDYIAAMSTQYESQ 307
A+ +D R A AD I A + E Q
Sbjct 264 --ASGKKIDNRVASETADSLIKAANASNELQ 292
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 79.3 bits (194), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 96/357 (27%), Positives = 150/357 (42%), Gaps = 55/357 (15%)
Query 48 DFNAREAQKARDFQ-----------LEMWNRQNEYNSPANQRKLRAEAGYNPY-LGYDSN 95
+FNA EA K RDFQ L+ WNR+N YN P+ QR AG+NPY + D+
Sbjct 47 EFNASEALKNRDFQTSEREASQQWNLDQWNRENAYNDPSAQRARMEAAGFNPYNMNIDAG 106
Query 96 TGVagstgstsqaqaasPLSLNPEV---------YSELGSQLGRAGQMIYQERESNARTK 146
+ S+ + + + S P + + + S + + G + +AR
Sbjct 107 SASTSGAQSSPGSGSQATASHTPSLPAYTGYAADFQNVASGIAQIGNAV--SSGIDARLT 164
Query 147 ALQGDADVARAQAL-QVFSNVDWGKLSPDYKKWMRETGLQRAQLDYDTNKQNLQNLRWSN 205
+ GD D+ +A + ++ N +W L+ YK + L D K+ L+NL
Sbjct 165 SAYGD-DLMKADIMSKIGGNSEW--LTDVYKLGRQNEAPNL--LGIDLRKKRLENLSTET 219
Query 206 LIQMAERTNLLLS--ANTKRTLNKYLDQSEQTRINVMAAQYYDLMAAGHLKYQQCKESIA 263
I++A LL A +R +NK++ +Q + A + AG L Q K I
Sbjct 220 NIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSEAQVKTQIK 279
Query 264 KQILYSKQGSWYDSMANKNNLDYRNALALADDYIAAMSTQYESQTAYNMGFGQKAQEAGR 323
+Q L Q ++ K L+ R A LAD AM+ +Y + AY GF A +AG
Sbjct 280 QQALLEAQ-----TVGQK--LNNRLAERLADYQFKAMAAEYRANAAYYNGFYNDAWQAGM 332
Query 324 RDAESKSFKSLIDR----------------WNYNKRYYEEGLNTLQVVANAIGSIRG 364
A ++S R W N YY G L+ + ++GS+ G
Sbjct 333 SKAAQARYESNAARIAADMSEIFKGREKSSWKNNPIYYNIG-ELLKGILGSVGSVAG 388
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 78.6 bits (192), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 97/357 (27%), Positives = 149/357 (42%), Gaps = 55/357 (15%)
Query 48 DFNAREAQKARDFQ-----------LEMWNRQNEYNSPANQRKLRAEAGYNPY-LGYDSN 95
+FNA EA K RDFQ L+ WNR+N YN P+ QR AG+NPY + D
Sbjct 47 EFNASEALKNRDFQTSEREASQQWNLDQWNRENAYNDPSAQRARMEAAGFNPYNMNIDPG 106
Query 96 TGVagstgstsqaqaasPLSLNPEV---------YSELGSQLGRAGQMIYQERESNARTK 146
+G S+ + +++ S P + + + S + + G + + AR
Sbjct 107 SGSTSGAQSSPGSGSSATASHTPSLPAYTGYAADFQNVASGIAQIGNAVASGID--ARLT 164
Query 147 ALQGDADVARAQAL-QVFSNVDWGKLSPDYKKWMRETGLQRAQLDYDTNKQNLQNLRWSN 205
+ GD D+ +A + ++ N +W L+ YK + L D K+ L+NL
Sbjct 165 SAYGD-DLMKADIMSKIGGNSEW--LTDVYKLGRQNEAPNL--LGIDLRKKRLENLSTET 219
Query 206 LIQMAERTNLLLS--ANTKRTLNKYLDQSEQTRINVMAAQYYDLMAAGHLKYQQCKESIA 263
I++A LL A +R +NK++ +Q + A + AG L Q K I
Sbjct 220 NIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSEAQVKTQIK 279
Query 264 KQILYSKQGSWYDSMANKNNLDYRNALALADDYIAAMSTQYESQTAYNMGFGQKAQEAGR 323
+Q L Q A L+ R A LAD AM+ +Y + AY GF A +AG
Sbjct 280 QQALLEAQ-------AAGQKLNNRLAERLADYQFKAMAAEYRANAAYYNGFYNDAWQAGM 332
Query 324 RDAESKSFKS------------LIDR----WNYNKRYYEEGLNTLQVVANAIGSIRG 364
A ++S DR W N YY L+ + ++GS+ G
Sbjct 333 SKAAQVRYESNAARIAAQMSEIFKDREKASWKNNPIYYNIS-ELLKGLLGSVGSVVG 388
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 77.8 bits (190), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 80/319 (25%), Positives = 148/319 (46%), Gaps = 52/319 (16%)
Query 53 EAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYLGYDSNTGVagstgstsqaqaas 112
E+QK+RDF M++ NE+NS +QR EAG NPYL + + + S + + +S
Sbjct 47 ESQKSRDFAKSMFDASNEWNSAKSQRARLEEAGLNPYLMMNGGSAGTAQSTSATASSGSS 106
Query 113 PLSLNPEVYSE---LGSQLGRAGQMIYQERESNARTKALQGDADVARAQALQVFSNVDWG 169
P Y+ +G AG M + S+AR + D L + + D+
Sbjct 107 GSGGMPYQYTPTNVIGDVASYAGAM---KSLSDARKSGTEADL-------LGRYGDSDYS 156
Query 170 KLSPDYKKWMRETGLQRAQLDYDTNKQNLQNLRWSNLIQMAERTNLLLSANTKRTLNKYL 229
+ + +T ++ Q D T A++ NLLLSA ++ +N YL
Sbjct 157 SRIANTEA---DTYFKQRQSDVAT----------------AQKANLLLSAEAQQVMNMYL 197
Query 230 DQSEQTRINVMAAQYYDLMAAGHLKYQQCKESIAKQILYSKQGSWYDSMANKNNLDYRNA 289
Q +Q +++ + AQY++++ G +K +Q K +A ++ ++ ++ + A
Sbjct 198 PQEKQIQLSTLGAQYWNMIRDGSIKEEQAKNLLATRLE-------IEARTAGQHISNKVA 250
Query 290 LALADDYIAAMSTQYESQTAYNMGFGQKAQEAGRRDAESKSFKSLIDRWNYN--KRYYEE 347
+ AD I A +T ++ AYN G+ Q + + G R + +DRW+ + K ++
Sbjct 251 RSTADSIIDATNTAKMNEAAYNRGYSQFSNDVGYRTGK-------MDRWSQDPVKARWDR 303
Query 348 GLNT----LQVVANAIGSI 362
G+N + ++N +GS+
Sbjct 304 GINNAGKFIDGLSNIVGSV 322
> Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.protein.BACEGG.02723
Length=354
Score = 56.2 bits (134), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 40/162 (25%), Positives = 83/162 (51%), Gaps = 20/162 (12%)
Query 207 IQMAERTNLLLSANTKRTLNKYLDQSEQTRINVMAAQYYDLMAAGHLKYQQCKESIAKQI 266
+ +A++ NLLL + + LN YL + ++ ++ + AQY++++ G + +Q K IA ++
Sbjct 175 VAIAQKANLLLRNDAQEVLNMYLPEEKRIQLQMNGAQYWNMIREGVISEEQAKNLIASRL 234
Query 267 LYSKQGSWYDSMANKNNLDYRNALALADDYIAAMSTQYESQTAYNMGFGQKAQEAGRRDA 326
++ ++ + A + AD I A T E++ A+N G+ Q + + G R
Sbjct 235 E-------IEARTQGQHISNKIAKSTADSIIDATRTAKENEAAFNRGYSQFSNDVGFRTG 287
Query 327 ESKSFKSLIDRW--NYNKRYYEEGLNT----LQVVANAIGSI 362
+ +DRW + K ++ G+N + ++NA+GS+
Sbjct 288 K-------MDRWLQDPVKARWDRGINNAGKFIDGLSNAVGSL 322
Score = 42.0 bits (97), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 20/38 (53%), Positives = 25/38 (66%), Gaps = 0/38 (0%)
Query 53 EAQKARDFQLEMWNRQNEYNSPANQRKLRAEAGYNPYL 90
E+QK+RDF M++ NE+NS NQR AG NPYL
Sbjct 47 ESQKSRDFAKSMFDATNEWNSAKNQRARLEAAGLNPYL 84
Lambda K H a alpha
0.313 0.127 0.365 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 31621345