bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
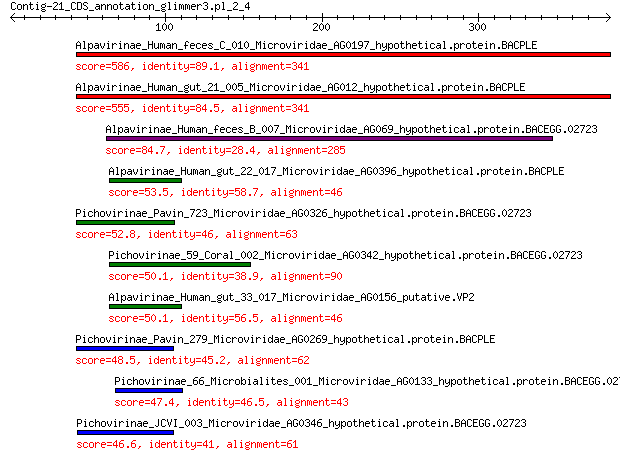
Query= Contig-21_CDS_annotation_glimmer3.pl_2_4
Length=383
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_C_010_Microviridae_AG0197_hypothetical.... 586 0.0
Alpavirinae_Human_gut_21_005_Microviridae_AG012_hypothetical.pr... 555 0.0
Alpavirinae_Human_feces_B_007_Microviridae_AG069_hypothetical.p... 84.7 6e-20
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 53.5 9e-10
Pichovirinae_Pavin_723_Microviridae_AG0326_hypothetical.protein... 52.8 1e-09
Pichovirinae_59_Coral_002_Microviridae_AG0342_hypothetical.prot... 50.1 6e-09
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 50.1 1e-08
Pichovirinae_Pavin_279_Microviridae_AG0269_hypothetical.protein... 48.5 3e-08
Pichovirinae_66_Microbialites_001_Microviridae_AG0133_hypotheti... 47.4 6e-08
Pichovirinae_JCVI_003_Microviridae_AG0346_hypothetical.protein.... 46.6 1e-07
> Alpavirinae_Human_feces_C_010_Microviridae_AG0197_hypothetical.protein.BACPLE
Length=382
Score = 586 bits (1510), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 304/341 (89%), Positives = 321/341 (94%), Gaps = 0/341 (0%)
Query 43 NKSNRKLSKEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLM 102
N+SNRK S EAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLM
Sbjct 42 NRSNRKASAEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLM 101
Query 103 YSRGDVGNatapeapqqaatpRYNVIPANTYGQTAQIAADAGLKAAQARLAsseskktdt 162
YSRGDVGNATAPEAP QA TPRYNVIP NTYGQTAQIAADAGLKAAQARLA+SESKKT+T
Sbjct 102 YSRGDVGNATAPEAPAQAPTPRYNVIPTNTYGQTAQIAADAGLKAAQARLANSESKKTET 161
Query 163 eesLLTADYLLRKARTESDIELNSSTIYVNHELGQLNHAEAEVAAKKLQEIDVAMSEARE 222
EE LLTADYLLRKARTESDIELN+STIYVNHELGQLNHAEAEVAAKKLQEIDVAMSEARE
Sbjct 162 EEGLLTADYLLRKARTESDIELNNSTIYVNHELGQLNHAEAEVAAKKLQEIDVAMSEARE 221
Query 223 RISTLKAQQSQIDENLVQMKFDRFLRSKEFELLCKKTYQDMKESNSRLTLNAAEVQDMMA 282
RI+T+KAQQSQIDEN+VQ+KFDR+LRS EFELLCKKTYQDMKESNSR+ LNAAEVQDMMA
Sbjct 222 RINTMKAQQSQIDENIVQLKFDRYLRSNEFELLCKKTYQDMKESNSRINLNAAEVQDMMA 281
Query 283 TQLARVMNLNASTYMQKKQGLLASEQTMTELYKQTGIDISNQHAKFNFDQAKNWDSTERF 342
TQLARVMNLNASTYMQKKQG+LASEQTMTELYKQTGIDISNQHAKFNFDQAKNWDSTERF
Sbjct 282 TQLARVMNLNASTYMQKKQGMLASEQTMTELYKQTGIDISNQHAKFNFDQAKNWDSTERF 341
Query 343 TNVATTWINSISCAVGQFTGSASDLKQAGFLSPPRNKAGFR 383
TNVATTWINS+S AV QF G+ + L++ GFL + GFR
Sbjct 342 TNVATTWINSVSFAVSQFAGATTSLQKGGFLGKSMSPIGFR 382
> Alpavirinae_Human_gut_21_005_Microviridae_AG012_hypothetical.protein.BACPLE
Length=383
Score = 555 bits (1431), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 288/341 (84%), Positives = 314/341 (92%), Gaps = 0/341 (0%)
Query 43 NKSNRKLSKEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLM 102
NKSNRK S EAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLM
Sbjct 43 NKSNRKQSAEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLM 102
Query 103 YSRGDVGNatapeapqqaatpRYNVIPANTYGQTAQIAADAGLKAAQARLAsseskktdt 162
YSRGDVGNATAPEAP QA TPR+NVIP NTYGQTAQIAAD GLKA+QARLA S+SKKTDT
Sbjct 103 YSRGDVGNATAPEAPAQAPTPRFNVIPTNTYGQTAQIAADTGLKASQARLADSQSKKTDT 162
Query 163 eesLLTADYLLRKARTESDIELNSSTIYVNHELGQLNHAEAEVAAKKLQEIDVAMSEARE 222
EESLLTADYLLRKART+S+I+LN+STIYVNHELGQLNHAEA++AAKKLQEIDVAMSEARE
Sbjct 163 EESLLTADYLLRKARTDSEIQLNNSTIYVNHELGQLNHAEADLAAKKLQEIDVAMSEARE 222
Query 223 RISTLKAQQSQIDENLVQMKFDRFLRSKEFELLCKKTYQDMKESNSRLTLNAAEVQDMMA 282
RI+TL+AQQS+IDE +VQMKFDR+LRSKEFELLC +TYQD+KESNSR++LNAAEVQD+MA
Sbjct 223 RINTLRAQQSEIDEKIVQMKFDRYLRSKEFELLCVRTYQDIKESNSRISLNAAEVQDIMA 282
Query 283 TQLARVMNLNASTYMQKKQGLLASEQTMTELYKQTGIDISNQHAKFNFDQAKNWDSTERF 342
TQLARV+NLNASTYM KKQGLLASEQTMTEL+KQTGIDISNQ A FNF QA+ WDSTERF
Sbjct 283 TQLARVLNLNASTYMMKKQGLLASEQTMTELFKQTGIDISNQQAIFNFQQAQTWDSTERF 342
Query 343 TNVATTWINSISCAVGQFTGSASDLKQAGFLSPPRNKAGFR 383
TNVATTWINS+S A GQF G+ + L + GFL P GFR
Sbjct 343 TNVATTWINSLSFAAGQFAGATTSLSKGGFLGKPMTPIGFR 383
> Alpavirinae_Human_feces_B_007_Microviridae_AG069_hypothetical.protein.BACEGG.02723
Length=383
Score = 84.7 bits (208), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 81/315 (26%), Positives = 152/315 (48%), Gaps = 43/315 (14%)
Query 62 REERLAQQQWIEQM--YEKNN---------SYNSPAAQMQRLKEAGLNPDLMYSRGDVGN 110
+EE + W E+M + NN SYNSP+ M RLK+AGLNPDL+Y G G
Sbjct 48 KEENAQARAWSEKMARWYANNERANLADERSYNSPSTVMSRLKDAGLNPDLIYGNGAAG- 106
Query 111 atapeapqqaatpRYNVIPANTYGQTAQIAADAGLKA-----------AQARLAsseskk 159
++ P +V PA+ G +A +++ A+ + +++ K
Sbjct 107 --LVDSNVAGTAPVSSVPPADVAGPI--MATPTAMESLFQGAAYAKTLAETKNIKADTSK 162
Query 160 tdteesLLTADYLLRKARTESDIELNSSTIYVNHELGQLNHAEAEVAAKKLQEIDVAMSE 219
+ E + L D ++ A +++ I+++ + QL A+AE K+ + +++
Sbjct 163 KEGEVTSLNIDNFVKAASSDNAIKMSGLEV-------QLTKAQAEYTEKQKSRLISEIND 215
Query 220 ARERISTLKAQQSQ-------IDENLVQMKFDRFLRSKEFELLCKKTYQDMKESNSRLTL 272
E ++ LKAQ S+ +D + V + L ++ F+L C++ + ++E+++++ L
Sbjct 216 INEHVNLLKAQISETWSRTANLDASTVATRTAAILNNRRFDLECEEFARRVRETDAKVNL 275
Query 273 NAAEVQDMMATQLARVMNLNASTYM-QKKQGLLASEQTMTELYKQTGIDISNQHAKFNFD 331
+ AE + ++ T A+V N++ T + Q L +++T E Y + IDI A F
Sbjct 276 SEAEAKSILVTMYAKVNNIDTDTALKQANIRLTDAQKTQVEHYTNS-IDIHRDAAVFKLQ 334
Query 332 QAKNWDSTERFTNVA 346
Q + +D +R VA
Sbjct 335 QDQKYDDAQRIVTVA 349
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 53.5 bits (127), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 27/48 (56%), Positives = 34/48 (71%), Gaps = 2/48 (4%)
Query 64 ERLAQQQ--WIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLMYSRGDVG 109
ER+A QQ W E M+ K N+YN+ +AQ QRL+EAGLNP LM + G G
Sbjct 47 ERMAIQQRNWQENMWNKENAYNTASAQRQRLEEAGLNPYLMMNGGSAG 94
> Pichovirinae_Pavin_723_Microviridae_AG0326_hypothetical.protein.BACEGG.02723
Length=266
Score = 52.8 bits (125), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/63 (46%), Positives = 40/63 (63%), Gaps = 11/63 (17%)
Query 43 NKSNRKLSKEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLM 102
N N+K S+E ++R+ R + L Q W +K N YNSP+ QMQR KEAGLNP+L+
Sbjct 28 NAQNKKFSQEMYDRQ----RADAL--QDW-----DKQNKYNSPSQQMQRYKEAGLNPNLI 76
Query 103 YSR 105
Y +
Sbjct 77 YGQ 79
> Pichovirinae_59_Coral_002_Microviridae_AG0342_hypothetical.protein.BACEGG.02723
Length=233
Score = 50.1 bits (118), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 35/105 (33%), Positives = 50/105 (48%), Gaps = 17/105 (16%)
Query 64 ERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLMYSRGDVGNatapeapqqaatp 123
+ LA +Q +E + N YN+P AQM+RLKEAGLNP+L+Y G N + +
Sbjct 27 QNLANRQNVE-FWNMQNKYNTPKAQMERLKEAGLNPNLIYGSGQT-NTGVAGSIAASKPA 84
Query 124 RYNV-------IPANTY--------GQTAQIAADAGLKAAQARLA 153
YN+ I A Y +T +AG+K + LA
Sbjct 85 PYNIQNPVPAAIAAGMYSPQKSLLEAKTVSTIKEAGIKGVELELA 129
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 50.1 bits (118), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 26/48 (54%), Positives = 34/48 (71%), Gaps = 2/48 (4%)
Query 64 ERLAQQQ--WIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLMYSRGDVG 109
ER+A QQ + E M+ K N+YN+ +AQ QRL+EAGLNP LM + G G
Sbjct 47 ERMAMQQRDFQENMWNKENTYNTASAQRQRLEEAGLNPYLMMNGGSSG 94
> Pichovirinae_Pavin_279_Microviridae_AG0269_hypothetical.protein.BACPLE
Length=292
Score = 48.5 bits (114), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 35/62 (56%), Gaps = 11/62 (18%)
Query 43 NKSNRKLSKEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLM 102
N+ NRK + + E ++A E+ AQ N YNSPAAQ QRLK+A LNP LM
Sbjct 45 NRKNRKFAIQIMNTERQWALEDAAAQ-----------NVYNSPAAQKQRLKDAELNPSLM 93
Query 103 YS 104
Y
Sbjct 94 YG 95
> Pichovirinae_66_Microbialites_001_Microviridae_AG0133_hypothetical.protein.BACEGG.02723
Length=275
Score = 47.4 bits (111), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 20/43 (47%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 68 QQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLMYSRGDVGN 110
Q+ + + + N YN P QMQR K+AGLNP L+Y +G+ GN
Sbjct 32 QRNFAVKFWNMQNEYNLPINQMQRFKDAGLNPHLIYGQGNAGN 74
> Pichovirinae_JCVI_003_Microviridae_AG0346_hypothetical.protein.BACEGG.02723
Length=252
Score = 46.6 bits (109), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/61 (41%), Positives = 33/61 (54%), Gaps = 11/61 (18%)
Query 44 KSNRKLSKEAFERESKFAREERLAQQQWIEQMYEKNNSYNSPAAQMQRLKEAGLNPDLMY 103
K R ++ +ER+ K A E+ +++ YNSP QMQRLKEAGLNP L Y
Sbjct 31 KEQRAYEEQIYERQRKDALED-----------WQRTTDYNSPEQQMQRLKEAGLNPKLAY 79
Query 104 S 104
Sbjct 80 G 80
Lambda K H a alpha
0.313 0.124 0.337 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 33550783