bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
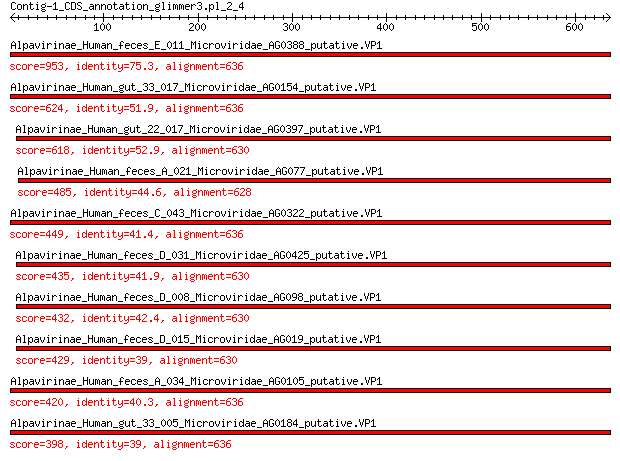
Query= Contig-1_CDS_annotation_glimmer3.pl_2_4
Length=636
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1 953 0.0
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 624 0.0
Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1 618 0.0
Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1 485 6e-165
Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1 449 2e-151
Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1 435 1e-145
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 432 2e-144
Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1 429 6e-144
Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1 420 6e-140
Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1 398 3e-131
> Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1
Length=622
Score = 953 bits (2463), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 479/637 (75%), Positives = 526/637 (83%), Gaps = 16/637 (3%)
Query 1 MAHFTGLKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRP 60
MAHFTGLKELQNHPHKAGFD+GSKNLFTAKVGEL+PVYWDFAIP CDYDIDLAYFTRTRP
Sbjct 1 MAHFTGLKELQNHPHKAGFDVGSKNLFTAKVGELIPVYWDFAIPDCDYDIDLAYFTRTRP 60
Query 61 VQTAAYTRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDPLTVGTDIPWCTL 120
VQTAAYTR+REYFDFYAVPCDLLWKSFDSAVIQMG+ APVQ+KT LDPLTVG DIPWCTL
Sbjct 61 VQTAAYTRVREYFDFYAVPCDLLWKSFDSAVIQMGQTAPVQSKTLLDPLTVGNDIPWCTL 120
Query 121 SDLYTSLIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGT 180
DL ++ F + LG TVSV + NIFGY+R D KLL YLNYGNFV PS SNVG+
Sbjct 121 LDLSNAVYFSSGSSPLGSTVSVPSGFGNIFGYNRGDVDSKLLFYLNYGNFVNPSLSNVGS 180
Query 181 STNRWFNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVD 240
+NRW+NTSF+SS V YSQKY N +V++FPLLAYQKIYQDFFRWSQWENADPT+YNVD
Sbjct 181 PSNRWWNTSFSSSKVPGYSQKYLNNNAVNIFPLLAYQKIYQDFFRWSQWENADPTSYNVD 240
Query 241 YYNGSGNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVVSG 300
YYNGSGNLFG+ G++ S+ +SN YWKRDNMFSLRYCNWNKDMF GLLPNSQFGDVAVV+
Sbjct 241 YYNGSGNLFGSSGLSGSVTASNSYWKRDNMFSLRYCNWNKDMFTGLLPNSQFGDVAVVNL 300
Query 301 VEGIDTFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSNTTPSRIRVAGGSGIPSSV 360
+ +PV + T T + +M++ + S + ++G + P S
Sbjct 301 GDSGSGTIPVGFLSD-----------TEVFTQAFNATSMSTVSDTSPMGISGST--PVSA 347
Query 361 AGSALG-VRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMSHM 419
S + + + FSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMSHM
Sbjct 348 RQSMVARINNADVASFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMSHM 407
Query 420 AQYIGGIARNLDISEVVNNNLSETGSEAVIYGKGVGTGSGKMRYHTGSQYCIIMCIYHAV 479
AQYIGG+ARNLDISEVVNNNL + GSEAVIYGKGVG+GSGKMRYHTGSQYCIIMCIYHA+
Sbjct 408 AQYIGGVARNLDISEVVNNNLKDDGSEAVIYGKGVGSGSGKMRYHTGSQYCIIMCIYHAM 467
Query 480 PLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNSNAFDNDLESDFDFLGYN 539
PLLDYAI+GQD QLLCTSVEDLPIPEFDNIGMEAVPA TLFNS FD + DFLGYN
Sbjct 468 PLLDYAITGQDPQLLCTSVEDLPIPEFDNIGMEAVPATTLFNSVLFDGTAVN--DFLGYN 525
Query 540 PRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWFASGGSSQASISWPFFKVNPNTLD 599
PRYWPWKSKIDRVHGAF TTLKDWVAPIDD YL+ WF S ASISWPFFKVNPNTLD
Sbjct 526 PRYWPWKSKIDRVHGAFTTTLKDWVAPIDDDYLHNWFNSKDGKSASISWPFFKVNPNTLD 585
Query 600 SIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
SIFAV ADS WE+DQLLINCDVSCKVVRPLSQDGMPY
Sbjct 586 SIFAVVADSIWETDQLLINCDVSCKVVRPLSQDGMPY 622
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 624 bits (1608), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 330/646 (51%), Positives = 435/646 (67%), Gaps = 42/646 (7%)
Query 1 MAHFTGLKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRP 60
MA +TGL LQNHPH++GFDIG KN FTAKVGELLPVYWD ++P Y ++ YFTRT+P
Sbjct 1 MAFYTGLSNLQNHPHRSGFDIGRKNAFTAKVGELLPVYWDISMPGDKYRFNVEYFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDPLTVGTDIPWCTL 120
V T+AYTR+REYFDFYAVP LLWKS S + QM +V +QA + L++GT +P T+
Sbjct 61 VATSAYTRLREYFDFYAVPLRLLWKSAPSVLTQMQDVNQIQALSLTQNLSLGTYLPSLTI 120
Query 121 SDLYTSLIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGT 180
L ++ ++N +T S + N FG+SR D S KLL YL YGN +E S +G
Sbjct 121 GTLGWAIRYLNGNTWEPETASYL---RNAFGFSRADLSFKLLSYLGYGNLIETPPS-LG- 175
Query 181 STNRWFNTSFTSS-AVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNV 239
NRW++TS ++ NY+Q+Y QN V++FPLL YQKIYQDFFRW QWE ++P++YNV
Sbjct 176 --NRWWSTSLKNTDDGANYTQQYIQNTIVNIFPLLTYQKIYQDFFRWPQWEKSNPSSYNV 233
Query 240 DYYNGSGNLFGNGGIASSIP-SSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVV 298
DY++GS + I S +P +S+ YWK D MF L+YCNWNKDM MG+LPNSQFGDVAV+
Sbjct 234 DYFSGS-----SPSIVSDLPAASSAYWKSDTMFDLKYCNWNKDMLMGVLPNSQFGDVAVL 288
Query 299 SGVEGIDTFVPVEVFNSINETNIAKPPLTGTH-TPVYTDDAMTSNTTPSRIRVAGGSGIP 357
D+ V + V + I + + P++ DA TSN P
Sbjct 289 DISSSGDSDVVLGVDPHKSTLGIGSAITSKSAVVPLFALDASTSN--------------P 334
Query 358 SSVAGSALGVRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMS 417
SV S + +F++LALRQAEALQ+WKEI+QS D++YR+QI HFG+ P ++S
Sbjct 335 VSVGSKLHLDLSSIKSQFNVLALRQAEALQRWKEISQSGDSDYREQILKHFGVKLPQALS 394
Query 418 HMAQYIGGIARNLDISEVVNNNLSETGSEAVIYGKGVGTGSGKMRYHTGSQYCIIMCIYH 477
++ YIGGI+RNLDISEVVNNNL+ AVI GKGVGTG+G Y T +++C+IM IYH
Sbjct 395 NLCTYIGGISRNLDISEVVNNNLAAEEDTAVIAGKGVGTGNGSFTYTT-NEHCVIMGIYH 453
Query 478 AVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNSNAFDNDLESDFDFL- 536
AVPLLDY ++GQD QLL T E LPIPEFDNIG+E +P +FNS+ L + F+
Sbjct 454 AVPLLDYTLTGQDGQLLVTDAESLPIPEFDNIGLEVLPMAQIFNSS-----LATAFNLFN 508
Query 537 -GYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWFASGGS-----SQASISWPF 590
GYNPRY+ WK+K+D ++GAF TTLK WV+P+ + L+ W G S ++A +++ F
Sbjct 509 AGYNPRYFNWKTKLDVINGAFTTTLKSWVSPVSESLLSGWARFGASDSKTGTKAVLNYKF 568
Query 591 FKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
FKVNP+ LD IF V ADSTW++DQLL+N + C VVR LS+DG+PY
Sbjct 569 FKVNPSVLDPIFGVKADSTWDTDQLLVNSYIGCYVVRNLSRDGVPY 614
> Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1
Length=607
Score = 618 bits (1594), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 333/657 (51%), Positives = 429/657 (65%), Gaps = 77/657 (12%)
Query 7 LKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRPVQTAAY 66
+ LQNHPH++GFDIG KN FTAKVGELLPVYWD ++P Y ++ YFTRT+PV+T+AY
Sbjct 1 MSNLQNHPHRSGFDIGRKNAFTAKVGELLPVYWDISMPGDKYKFNVEYFTRTQPVETSAY 60
Query 67 TRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDPLTVGTDIPWCTLSDLYTS 126
TR+REYFDFYAVP LLWKS S + QM ++ +QA + L++G+ P TLS
Sbjct 61 TRLREYFDFYAVPLRLLWKSAPSVLTQMQDINQLQALSFTQNLSLGSYFPSLTLSRFTAV 120
Query 127 LIFMNNRVSLGQTV-SVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGTSTNRW 185
L NR++ G V + N FG+SR D + KL YL YGN S S+NRW
Sbjct 121 L----NRLNGGSNVPGNSSTFLNEFGFSRADLAFKLFSYLGYGNVWSSEFS----SSNRW 172
Query 186 FNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVDYYNGS 245
++TS +Y+Q+Y Q+ V+LFPLLAYQKIYQDFFRWSQWEN++P++YNVDY+ G
Sbjct 173 WSTSLKGGG--SYTQQYVQDSYVNLFPLLAYQKIYQDFFRWSQWENSNPSSYNVDYFTGV 230
Query 246 GNLFGNGGIASSIP-SSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVA-------- 296
L + SS+P +SN YWK MF L+YCNWNKDM MG+LPNSQFGDVA
Sbjct 231 SPL-----LVSSLPEASNAYWKSPTMFDLKYCNWNKDMLMGVLPNSQFGDVAVLDIDNSG 285
Query 297 ---VVSGVEGIDTFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSNTTPSRIRVAGG 353
VV G+ ++ V V + S N +I P + A ++NT P
Sbjct 286 KPDVVLGLGNANSTVGVASYVSSNTASI----------PFFALKASSANTLP-------- 327
Query 354 SGIPSSVAGSALGVR-SLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINT 412
GS L V + L +F++LALRQAEALQ+WKEI+QS D++YR+QI+ HFG+N
Sbjct 328 -------VGSTLRVDLASLKSQFTVLALRQAEALQRWKEISQSGDSDYREQIRKHFGVNL 380
Query 413 PASMSHMAQYIGGIARNLDISEVVNNNLSETGSEAVIYGKGVGTGSGKMRYHTGSQYCII 472
P ++S+M YIGGI+RNLDISEVVNNNL+ G AVI GKGVG G+G Y T +++C++
Sbjct 381 PQALSNMCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAGNGSFTYTT-NEHCVV 439
Query 473 MCIYHAVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNS------NAFD 526
MCIYHAVPLLDY I+GQD QLL T E LPIPEFDNIGME +P +FNS N F+
Sbjct 440 MCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMETLPMTQIFNSPKASIVNLFN 499
Query 527 NDLESDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWFASGGS----- 581
GYNPRY+ WK+K+D ++GAF TTLK WV+P+ + L+ WF G S
Sbjct 500 ---------AGYNPRYFNWKTKLDVINGAFTTTLKSWVSPVTESLLSGWFGFGYSEGDVN 550
Query 582 --SQASISWPFFKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
++ +++ FFKVNP+ LD IF VAADSTW+SDQLL+N + C V R LS+DG+PY
Sbjct 551 SQNKVVLNYKFFKVNPSVLDPIFGVAADSTWDSDQLLVNSYIGCYVARNLSRDGVPY 607
> Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1
Length=622
Score = 485 bits (1248), Expect = 6e-165, Method: Compositional matrix adjust.
Identities = 280/646 (43%), Positives = 375/646 (58%), Gaps = 49/646 (8%)
Query 9 ELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRPVQTAAYTR 68
+L+NHP ++GFD+ + FT+K GELLPVYW IP ++I +FTRT+PV TAAYTR
Sbjct 8 DLKNHPRRSGFDLTKRICFTSKAGELLPVYWKPTIPGDKWNISTNWFTRTQPVDTAAYTR 67
Query 69 IREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDPLTVGTDIPWCTLSDLYTSLI 128
++EY D++ VP +L+ K +SA+ QM + PV A + ++ + TD+P+ TL L +L
Sbjct 68 VKEYVDWFFVPLNLIQKGIESAITQMVD-NPVSAMSAIENRAITTDMPYTTLLSLSRALY 126
Query 129 FMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGTSTNRWFNT 188
+N + + + N+FG+SR D S KLL L YGNF+ P S + T
Sbjct 127 MLNGKSYVNSHAGKL----NMFGFSRADLSAKLLQMLKYGNFINPEHSGLDTPM------ 176
Query 189 SFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVDYYNGSGNL 248
F S V+ Y N +V++ P+ YQKIY D+FR+ QWE P YN DYY G G++
Sbjct 177 -FGYSTVKLAQFSYLWNQAVNVLPIFCYQKIYSDYFRFQQWEKPVPYTYNADYYAG-GDI 234
Query 249 FGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVVSGVEGIDTFV 308
F N +++P + YW F LRYCNWN+D++ G LPN QFG+V+VV D +
Sbjct 235 FPN---PTTMPDA--YWNNYTPFDLRYCNWNRDLYTGFLPNQQFGNVSVVDMTVSSDEIM 289
Query 309 --PVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSNTTPSRIRVAGGSGIPSSVAGSALG 366
PV + N + K L + TS+T + G +P + A
Sbjct 290 AAPVTFGATGNRVAVTKAALASS----------TSSTGIGTVSTPSGGTVPVGTSLYAQV 339
Query 367 VRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMSHMAQYIGGI 426
+ L G FSILALRQAE LQKWKEI S D +YR QI+ HFG+ PA +S+M+QYIGG
Sbjct 340 QQRDLTGAFSILALRQAEFLQKWKEIALSGDQDYRSQIEKHFGVKLPAELSYMSQYIGGQ 399
Query 427 ARNLDISEVVNNNLS-ETGSEAVIY-----GKGVGTGSGKMRYHTGSQYCIIMCIYHAVP 480
+DISEVVN NL+ + GS+A Y GKGV +G G + Y T Q+ IIM IYHAVP
Sbjct 400 FAQMDISEVVNQNLTDQAGSDAAQYPALIAGKGVNSGDGNVNY-TARQHGIIMGIYHAVP 458
Query 481 LLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNSNAFDNDLESDFDF----- 535
LLDY +GQD LL TS E+ IPEFD IGM+ +P TLFNSN D SDF
Sbjct 459 LLDYERTGQDQDLLITSAEEWAIPEFDAIGMQTLPLGTLFNSNKVSGD--SDFRLHGAAY 516
Query 536 -LGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWFA----SGGSSQASISWPF 590
+GY PRY WK+ ID + GAF ++ K WVAPID ++ W + + Q+ ++ +
Sbjct 517 PIGYVPRYVNWKTDIDEIFGAFRSSEKTWVAPIDADFITNWVKNVADNASAVQSLFNYNW 576
Query 591 FKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
FKVNP LD IFAV ADST ++D +S K VR L GMPY
Sbjct 577 FKVNPAILDDIFAVKADSTMDTDTFKTRMMMSIKCVRNLDYSGMPY 622
> Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1
Length=602
Score = 449 bits (1156), Expect = 2e-151, Method: Compositional matrix adjust.
Identities = 263/645 (41%), Positives = 373/645 (58%), Gaps = 52/645 (8%)
Query 1 MAHFTGLKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRP 60
MA+ ++N P ++GFD FTAK GELLPVYW +P +++L+ FTRT P
Sbjct 1 MANLFSYGSVKNKPARSGFDWSEHFSFTAKAGELLPVYWKMLLPGTKVNLNLSSFTRTMP 60
Query 61 VQTAAYTRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDPLTVGTDIPWCTL 120
V TAAYTR++EY+D+Y VP L+ KS A++QM + PVQA + + +V D+PW
Sbjct 61 VNTAAYTRVKEYYDWYFVPLRLINKSIGQALVQMQD-QPVQATSIVANKSVTLDLPWTNA 119
Query 121 SDLYTSLIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGN-FVEPSSSNVG 179
+ ++T L + N ++ + N+ G+S+ TS KLL YL YGN + S VG
Sbjct 120 ATMFTLLNYAN---------VILTNKYNLDGFSKAATSAKLLRYLRYGNCYYTSDPSKVG 170
Query 180 TSTNRWFNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNV 239
N +F S+ +++ ++NVS ++ PL AYQKIY D+FR+ QWENA P YN
Sbjct 171 K------NKNFGLSSKDDFNLLAAKNVSFNVLPLAAYQKIYCDWFRFEQWENACPYTYNF 224
Query 240 DYYNGSGNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVVS 299
DYYNG GN+F A + ++W DN+ SLRY N+NKD+FMG++P+SQFG VA V+
Sbjct 225 DYYNG-GNVF-----AGVTANPENFWSNDNILSLRYANYNKDLFMGVMPSSQFGSVATVN 278
Query 300 GVEGIDTFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSNTTPSRIRVAGGSGIPSS 359
V F+S +N++ P ++ + T S+ +P +R + +
Sbjct 279 ----------VANFSS---SNLSSPLRNLSNVGMVTASNNASSGSPLILRSSQDTS---- 321
Query 360 VAGSALGVR-----SLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPA 414
AG G+ S L F IL+ R AEA QKWKE+TQ Y++Q++AHF +
Sbjct 322 -AGDGFGILTSNIFSTLSASFDILSFRIAEATQKWKEVTQCAKQGYKEQLEAHFNVKLSE 380
Query 415 SMSHMAQYIGGIARNLDISEVVNNNLSETGSEAVIYGKGVGTGSGKMRYHTGSQYCIIMC 474
++S +YIGG + + ISEV+N NL ++ I GKGVG G + T +++ I+MC
Sbjct 381 ALSDHCRYIGGTSSGVTISEVLNTNLESAAAD--IKGKGVGGSFGSETFET-NEHGILMC 437
Query 475 IYHAVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNSNAFD-NDLESDF 533
IYHA P+LDY SGQD QLL T D+PIPEFD+IGMEA+P TLFN + + L +
Sbjct 438 IYHATPVLDYLRSGQDLQLLSTLATDIPIPEFDHIGMEALPIETLFNEQSTEATALINSI 497
Query 534 DFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAP--IDDFYLNRWFASGGSSQASISWPFF 591
LGY+PRY +K+ +D V GAF TTL WVAP +++ F S S+++ FF
Sbjct 498 PVLGYSPRYIAYKTSVDWVSGAFETTLDSWVAPLTVNEQITKLLFNPDSGSVYSMNYGFF 557
Query 592 KVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
KV P LD IF TW+SDQ L+N + K V+ L +GMPY
Sbjct 558 KVTPRVLDPIFVQECTDTWDSDQFLVNVSFNVKPVQNLDYNGMPY 602
> Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1
Length=614
Score = 435 bits (1118), Expect = 1e-145, Method: Compositional matrix adjust.
Identities = 264/650 (41%), Positives = 375/650 (58%), Gaps = 56/650 (9%)
Query 7 LKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRPVQTAAY 66
+ ++NHP ++GFD+ ++ FT+K GELLPV+WD P + I FTRT+P+ TAAY
Sbjct 1 MSAVKNHPRRSGFDLSNRVCFTSKAGELLPVFWDIVYPGDSFKIKTQLFTRTQPLNTAAY 60
Query 67 TRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDPLTVGTDIPWC--TLSDLY 124
TRIREY DFY VP L+ K+ +A++QM + PVQA V TDIPW LS Y
Sbjct 61 TRIREYLDFYFVPLRLINKNLPTALMQMQD-NPVQATGLSSNKVVTTDIPWVPTNLSGTY 119
Query 125 TSLIFMNN-RVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGTSTN 183
SL + + + S + + + D + G+ S KLL+YL YGNF+ S+V + N
Sbjct 120 GSLTALADVKNSFPSSSTGIED---LLGFDAITQSAKLLMYLRYGNFL----SSVVSDKN 172
Query 184 RWFNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVDYYN 243
+ S S ++N + S+ + PL AYQK Y DFFR++QWE P YN D+Y+
Sbjct 173 KSLGLS-GSLDLRNSETASTGYTSMHILPLAAYQKAYADFFRFTQWEKNQPYTYNFDWYS 231
Query 244 GSGNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVV--SGV 301
G GN+ + +++ + Y+ DN+F+LRY NW KDMFMG++P+SQ GDV++V SG
Sbjct 232 G-GNVLAS---LTTLDLAKKYYSDDNLFTLRYANWPKDMFMGVMPDSQLGDVSIVDASGS 287
Query 302 EGIDTFVPVEVFNSINETNI--------AKPPLTGTHTPVYTDDAMTSNTTPSRIRVAGG 353
EG TF PV + + +N+ + P + + T A+++NTT +
Sbjct 288 EG--TF-PVGLLD-VNDGTLRAGLLARSGSAPAEKSSLEMQTSSALSANTTYG-VYAQRA 342
Query 354 SGIPSSVAGSALGVRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTP 413
+G+ SS FSIL LR AEA+QK++E++Q D + R QI AHFG++
Sbjct 343 AGLASS---------------FSILQLRMAEAVQKYREVSQFADQDARGQIMAHFGVSLS 387
Query 414 ASMSHMAQYIGGIARNLDISEVVNNNLSETGSE-AVIYGKGVGTGSGKMRYHTGSQYCII 472
+S Y+GG + N+D+SEVVN N+ TG A I GKGVGTG G +Y II
Sbjct 388 PVLSDKCVYLGGSSSNIDLSEVVNTNI--TGDNVAEIAGKGVGTGQGSFSGQF-DEYGII 444
Query 473 MCIYHAVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNSNAFD--NDLE 530
+ IYH VPLLDY I+GQ LL T+ DLP PEFD+IGM+ + NS + + ++
Sbjct 445 IGIYHNVPLLDYVITGQPQNLLYTNTADLPFPEFDSIGMQTIQFGRFVNSKSVSWTSGVD 504
Query 531 SDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWFASGGSSQASIS--- 587
+GY PR++ K++ D V GAF +TLK+WVAP+D Y+++W S +S ++
Sbjct 505 YRVQTMGYLPRFFDVKTRYDEVLGAFRSTLKNWVAPLDPSYVSKWLQSSVTSSGKLALNL 564
Query 588 -WPFFKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
+ FFKVNP LDSIF V DST ++DQ L + K VR DGMPY
Sbjct 565 NYGFFKVNPRVLDSIFNVKCDSTIDTDQFLTALYMDIKAVRNFDYDGMPY 614
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 432 bits (1110), Expect = 2e-144, Method: Compositional matrix adjust.
Identities = 267/651 (41%), Positives = 367/651 (56%), Gaps = 59/651 (9%)
Query 7 LKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRPVQTAAY 66
+ ++NHP ++GFD+ ++ FT+K GELLPV+WD P + I FTRT+P+ TAAY
Sbjct 6 MSAVKNHPRRSGFDLSNRVCFTSKAGELLPVFWDIVYPGDSFKIKTQLFTRTQPLNTAAY 65
Query 67 TRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDPLTVGTDIPWCTLSDLYTS 126
TRIREY DFY VP L+ K+ +A+ QM + PVQA V TDIPW + S
Sbjct 66 TRIREYLDFYFVPLRLINKNLPTALTQMQD-NPVQATGLSSNKVVTTDIPWVPVHSSSGS 124
Query 127 LIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNF----VEPSSSNVGTST 182
+ + +VS D N G+ S KLL+YL YGNF V ++G S+
Sbjct 125 YSSLTGFADVRGSVS-SSDIENFLGFDSITQSAKLLMYLRYGNFLSSVVPDEQKSIGLSS 183
Query 183 NRWFNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVDYY 242
+ S T S + SV + PL YQKIY DFFR++QWE P YN D+Y
Sbjct 184 SLDLRNSETVS---------TGYTSVHILPLATYQKIYADFFRFTQWEKNQPYTYNFDWY 234
Query 243 NGSGNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVV--SG 300
+G GN+ + ++ + Y+ DN+F+LRY NW KDMFMG++P+SQ GDV++V SG
Sbjct 235 SG-GNVLAS---LTTQALAKKYYSDDNLFTLRYANWPKDMFMGVMPDSQLGDVSIVDTSG 290
Query 301 VEGIDTFVPVEVFNSINETNIA-------KPPLTGTHTPVYTDDAMTSNTTPSRIRVAGG 353
EG TF PV ++N + + A P G+ + T A++++T
Sbjct 291 SEG--TF-PVGLYNFADGGSRAGLVAVSGTSPAAGSSLDMQTTSALSASTK--------- 338
Query 354 SGI-PSSVAGSALGVRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINT 412
G+ VAG LG FSIL LR AEA+QK++E++Q D + R QI AHFG++
Sbjct 339 YGVYAQQVAG--------LGSSFSILQLRMAEAVQKYREVSQFADQDARGQIMAHFGVSL 390
Query 413 PASMSHMAQYIGGIARNLDISEVVNNNLSETGSE-AVIYGKGVGTGSGKMRYHTGSQYCI 471
+S Y+GG + N+D+SEVVN N+ TG A I GKGVGTG G + + Y I
Sbjct 391 SPVLSDKCMYLGGSSSNIDLSEVVNTNI--TGDNIAEIAGKGVGTGQGSFSGNFDT-YGI 447
Query 472 IMCIYHAVPLLDYAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNSNA--FDNDL 529
IM IYH VPLLDY I+GQ LL T+ DLP PE+D+IGM+ + NS A + + +
Sbjct 448 IMGIYHNVPLLDYVITGQPQNLLYTNTADLPFPEYDSIGMQTIQFGRFVNSKAVGWTSGV 507
Query 530 ESDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWF----ASGGSSQAS 585
+ +GY PR++ K++ D V GAF +TLK+WVAP+D L +W S G +
Sbjct 508 DYRTQTMGYLPRFFDVKTRYDEVLGAFRSTLKNWVAPLDPANLPQWLQTSVTSSGKLFLN 567
Query 586 ISWPFFKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
+++ FFKVNP LDSIF V DST ++DQ L + K VR DGMPY
Sbjct 568 LNYGFFKVNPRVLDSIFNVKCDSTIDTDQFLTTLYMDIKAVRNFDYDGMPY 618
> Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1
Length=584
Score = 429 bits (1104), Expect = 6e-144, Method: Compositional matrix adjust.
Identities = 246/636 (39%), Positives = 364/636 (57%), Gaps = 63/636 (10%)
Query 7 LKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRPVQTAAY 66
L ++N P ++GFD+ SK F+AKVGELLP+ W +P + + +FTRT+PV T+AY
Sbjct 6 LSSVKNRPRRSGFDLSSKVAFSAKVGELLPIKWTLTMPGDKFSLKEQHFTRTQPVNTSAY 65
Query 67 TRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLD-PLTVGTDIPWCTLSDLYT 125
TR+REY+D++ P LLW++ + Q+ + VQ + D + +G+++P + +
Sbjct 66 TRVREYYDWFWCPLHLLWRNAPEVIAQIQQ--NVQHASSFDGSVLLGSNMPCFSADQISQ 123
Query 126 SLIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGTSTNRW 185
SL M +++ N FG++R D ++KL+ YL YGN + VGTS +R
Sbjct 124 SLDMMKSKL-------------NYFGFNRADLAYKLIQYLRYGNV----RTGVGTSGSRN 166
Query 186 FNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVDYYNGS 245
+ TS +Y+Q + N ++S+FP+LAY+K QD+FR +QW+ + P +N+DYY+G
Sbjct 167 YGTSVDVKD-SSYNQNRAFNHALSVFPILAYKKFCQDYFRLTQWQVSAPYLWNIDYYDGK 225
Query 246 GNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVVSGVEGID 305
G + + + S Y++ D F L YCNWNKDMF G LP++Q+GD +VV G
Sbjct 226 G---ATTILPADLSKSVTYFEHDTFFDLEYCNWNKDMFFGSLPDAQYGDTSVVDISYGT- 281
Query 306 TFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSNTTPSRIRVAGGSGIPSSVAGSAL 365
T PV I N+ P + T + T D ++ AG+ L
Sbjct 282 TGAPV-----ITAQNLQSP--VNSSTAIGTSDKFSTQLIE---------------AGTNL 319
Query 366 GVRSLLGGEFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMSHMAQYIGG 425
+LALR+ EALQ+++EI+ NYR QIKAHFG++ + +S M+ YIGG
Sbjct 320 --------TLDVLALRRGEALQRFREISLCTPLNYRSQIKAHFGVDVGSELSGMSTYIGG 371
Query 426 IARNLDISEVVNNNLSETGSEAVIYGKGVGTGSGKMRYHTGSQYCIIMCIYHAVPLLDYA 485
A +LDISEVVN N++E+ +EA+I GKG+GTG G ++ + ++MCIYH+VPLLDY
Sbjct 372 EASSLDISEVVNTNITES-NEALIAGKGIGTGQGNEEFY-AKDWGVLMCIYHSVPLLDYV 429
Query 486 ISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNSNAFD-----NDLESDFDFLGYNP 540
IS D QL + P+PE D IG+E + + +++N + ++ +GY P
Sbjct 430 ISAPDPQLFASMNTSFPVPELDAIGLEPI-TVAYYSNNPIELPSTGGITDAPTTTVGYLP 488
Query 541 RYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWFASGGSSQASISWPFFKVNPNTLDS 600
RY+ WK+ ID V GAF TT K+WVAPI + G+ I++ FFKVNP+ LD
Sbjct 489 RYYAWKTSIDYVLGAFTTTEKEWVAPITPELWSNMLKPLGTKGTGINYNFFKVNPSILDP 548
Query 601 IFAVAADSTWESDQLLINCDVSCKVVRPLSQDGMPY 636
IFAV ADS W++D LIN +V R L DGMPY
Sbjct 549 IFAVNADSYWDTDTFLINAAFDIRVARNLDYDGMPY 584
> Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1
Length=618
Score = 420 bits (1080), Expect = 6e-140, Method: Compositional matrix adjust.
Identities = 256/667 (38%), Positives = 358/667 (54%), Gaps = 80/667 (12%)
Query 1 MAHFTGLKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRP 60
M+ +++N P ++GFD+ +K FTAKVGELLPVYW F +P ++I +FTRT+P
Sbjct 1 MSSLFSYGDIKNSPRRSGFDLSNKCAFTAKVGELLPVYWKFCLPGDKFNISQEWFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDPLTVGTDIPWCTL 120
V T+A+TRIREY++++ VP LL+++ + A++ M E P A + ++ ++PW L
Sbjct 61 VDTSAFTRIREYYEWFFVPLHLLYRNSNEAIMSM-ENQPNYAASGSSSISFNRNLPWVDL 119
Query 121 SDLYTSLIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGT 180
S + V++G S N FG SR + KL+ YL YG
Sbjct 120 STI---------NVAIGNVQSSTSP-KNFFGVSRSEGFKKLVSYLGYG------------ 157
Query 181 STNRWFNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVD 240
+ +KY N+ S FPL AYQKIYQD++R SQWE + P YN D
Sbjct 158 ---------------ETSPEKYVDNLRCSAFPLYAYQKIYQDYYRHSQWEKSKPWTYNCD 202
Query 241 YYNGSGNLFGNGGIASSIPSSNDYWKRDNMFSLRYCNWNKDMFMGLLPNSQFGDVAVVS- 299
++NG + + S ND ++F LRY NWNKD++MG LPNSQFGDVA +S
Sbjct 203 FWNGEDSTPITATVELFSQSPND-----SVFELRYANWNKDLWMGSLPNSQFGDVAGISL 257
Query 300 ---------GVEGIDTFVPVEVFNSINETNIAKPPLTGTHTPVYTDDAMTSNTTPSRI-- 348
GV G + + + + PV +DA T +
Sbjct 258 GFDASTMKVGVTGTALVKGNMPVGYGGKDGMGIRSQSRLYNPVGINDAQQVTTVQQDVNN 317
Query 349 ---------------RVAGGSGIPSSVAGSALGVRSLLGGEFSILALRQAEALQKWKEIT 393
R++ + I S + L + L +FS+L LR AEALQKWKEI
Sbjct 318 KENGYLFATGTDAFGRISNAAKINGSELFAQLSGQ--LDAQFSVLQLRAAEALQKWKEIA 375
Query 394 QSVDTNYRDQIKAHFGINTPASMSHMAQYIGGIARNLDISEVVNNNLSETGSEAVIYGKG 453
Q+ NY Q+KAHFG++T SH + I G ++DIS V N NL T EA+I GKG
Sbjct 376 QANGQNYAAQVKAHFGVSTNPMQSHRSTRICGFDGSIDISAVENTNL--TADEAIIRGKG 433
Query 454 VGTGS-GKMRYHTGSQYCIIMCIYHAVPLLDYAISGQDSQLLCT-SVEDLPIPEFDNIGM 511
+G T +++ +IMCIYHA PLLDY +G D QL+ T E P+PEFD++GM
Sbjct 434 LGGQRINDPSDFTCNEHGVIMCIYHATPLLDYVPTGPDLQLMSTVKGESWPVPEFDSLGM 493
Query 512 EAVPAITLFNSNAFDNDLESDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFY 571
E++P ++L NS A + + + GY PRY WK+ ID V GAF TLK W AP+D Y
Sbjct 494 ESLPMLSLVNSKAIGDIVAR--SYAGYVPRYISWKTSIDVVRGAFTDTLKSWTAPVDSDY 551
Query 572 LNRWFASGGSSQAS--ISWPFFKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVVRPL 629
++ +F + S +S+ +FKVNP+ L+ IFAV+ D +W +DQLL NC KV R L
Sbjct 552 MHVFFGEVIPQEGSPILSYTWFKVNPSVLNPIFAVSVDGSWNTDQLLCNCQFDVKVARNL 611
Query 630 SQDGMPY 636
S DGMPY
Sbjct 612 SYDGMPY 618
> Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1
Length=618
Score = 398 bits (1022), Expect = 3e-131, Method: Compositional matrix adjust.
Identities = 248/670 (37%), Positives = 353/670 (53%), Gaps = 86/670 (13%)
Query 1 MAHFTGLKELQNHPHKAGFDIGSKNLFTAKVGELLPVYWDFAIPSCDYDIDLAYFTRTRP 60
MA +++N P ++GFD+G+KN FTAKVGELLPVYW F +P + I +FTRT+P
Sbjct 1 MAGLFSYGDIKNKPRRSGFDLGNKNAFTAKVGELLPVYWKFCMPGDKFHISQEWFTRTQP 60
Query 61 VQTAAYTRIREYFDFYAVPCDLLWKSFDSAVIQMGEVAPVQAKTPLDPLTVGTDIPWCTL 120
V T+A+TRIREY++++ VP L++++ + A++ + E P A + + +PW L
Sbjct 61 VDTSAFTRIREYYEWFFVPLHLMYRNSNEAIMSL-ENQPNYAASGTQSIVFNRKLPWVDL 119
Query 121 SDLYTSLIFMNNRVSLGQTVSVVPDYSNIFGYSRCDTSHKLLLYLNYGNFVEPSSSNVGT 180
L ++ V ++N+FG++R +KL L G +PS
Sbjct 120 QTLNDAIT----------NVQASTYHNNMFGFARSSGFYKLFNSLGVGE-TDPS------ 162
Query 181 STNRWFNTSFTSSAVQNYSQKYSQNVSVSLFPLLAYQKIYQDFFRWSQWENADPTAYNVD 240
K N+ +S FP AYQKIY D +R SQWE P YN D
Sbjct 163 --------------------KTLANLRISAFPFYAYQKIYSDHYRNSQWEVNKPWTYNCD 202
Query 241 YYNGSGNLFGNGGIASSIPSSNDYWK---RDNMFSLRYCNWNKDMFMGLLPNSQFGDVAV 297
++NG ++ + + D + D++F LRY NWNKD++MG +PN+QFGDVA
Sbjct 203 FWNGED--------STPVAFTKDLFDTNPNDSVFELRYANWNKDLYMGAMPNTQFGDVAA 254
Query 298 VS-GVE------GIDTFVPVEVFNSIN---ETNIAKPPLTGTHTPVYTDDAMTSNTTPSR 347
VS G + GI PV + + + + + PV +DA T
Sbjct 255 VSLGFDTSTMKIGITGTAPVTGNMPVGYGGKDGMGLRSQSRLYNPVGINDAQQVTTVQEE 314
Query 348 IR-------VAGGSGI------PSSVAGSALGVR--SLLGGEFSILALRQAEALQKWKEI 392
+ A G+ + ++GS L + L +FS+L LR AE LQKWKEI
Sbjct 315 VNNKENGYLFATGTNAFGRIFNAAKISGSDLNAQLSGQLDAKFSVLQLRAAECLQKWKEI 374
Query 393 TQSVDTNYRDQIKAHFGINTPASMSHMAQYIGGIARNLDISEVVNNNLSETGSEAVIYGK 452
Q+ NY Q+KAHFG++ SH + + G ++DIS V N NLS EA+I GK
Sbjct 375 AQANGQNYAAQVKAHFGVSPNPITSHRSTRVCGFDGSIDISAVENTNLSS--DEAIIRGK 432
Query 453 GVGT-GSGKMRYHTGSQYCIIMCIYHAVPLLDYAISGQDSQLLCTSVED-LPIPEFDNIG 510
G+G K +++ ++MCIYHAVPLLDYA +G D Q + T D P+PE D+IG
Sbjct 433 GIGGYRVNKPETFETTEHGVLMCIYHAVPLLDYAPTGPDLQFMTTVDGDSWPVPEMDSIG 492
Query 511 MEAVPAITLFNSNAFDNDLESDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDF 570
E +P+ +L N+NA E GY PRY WK+ +D V GAF+ TLK W API
Sbjct 493 FEELPSYSLLNTNAVQPIKEP--RPFGYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGQD 550
Query 571 YLNRWF----ASGGSSQASISWPFFKVNPNTLDSIFAVAADSTWESDQLLINCDVSCKVV 626
YL +F GG+ +W FKVNP+ ++ IF V AD +W +DQLL+NCD +V
Sbjct 551 YLKIYFDNNNVPGGAHFGFYTW--FKVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVA 608
Query 627 RPLSQDGMPY 636
R LS DG+PY
Sbjct 609 RNLSYDGLPY 618
Lambda K H a alpha
0.319 0.135 0.419 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 58948742