bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
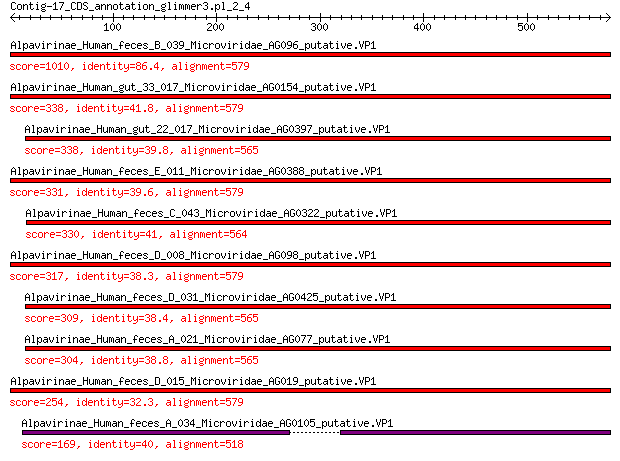
Query= Contig-17_CDS_annotation_glimmer3.pl_2_4
Length=579
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_B_039_Microviridae_AG096_putative.VP1 1010 0.0
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 338 3e-109
Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1 338 5e-109
Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1 331 2e-106
Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1 330 4e-106
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 317 6e-101
Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1 309 6e-98
Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1 304 5e-96
Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1 254 1e-77
Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1 169 6e-47
> Alpavirinae_Human_feces_B_039_Microviridae_AG096_putative.VP1
Length=579
Score = 1010 bits (2611), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 500/579 (86%), Positives = 540/579 (93%), Gaps = 0/579 (0%)
Query 1 MSDFNPLNRARISTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVP 60
MSDFNPL+RA+I THRSSFDLSSKKLFTAKVGEILPCYWQIAIPG KYRISSDWFTRTVP
Sbjct 1 MSDFNPLDRAKIPTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGTKYRISSDWFTRTVP 60
Query 61 VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQT 120
VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDY+TSAASSTAN+S LTSVP V+Q+
Sbjct 61 VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYMTSAASSTANTSALTSVPSVTQS 120
Query 121 LFNAFFQTANAGDQPNTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKKYLGVD 180
LF+AFFQTANAGDQPNTRDDAGLPIVYGSCKLLDMLGYGSMI S N KAAITKKYLGVD
Sbjct 121 LFSAFFQTANAGDQPNTRDDAGLPIVYGSCKLLDMLGYGSMIDSKNTGKAAITKKYLGVD 180
Query 181 SLSDADNPLVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGAGNIGLV 240
+L DADNPLVYQ+SQTVNALPFLAYQKIYYDFFSN+QWEKHKAYAYNVDYWSG GNIGLV
Sbjct 181 ALGDADNPLVYQSSQTVNALPFLAYQKIYYDFFSNNQWEKHKAYAYNVDYWSGTGNIGLV 240
Query 241 TDMVQLRYANYPKDYFMGMLPSSQYGSVAVLPsissssdsrsLLYYEPsssataalqsag 300
TDMVQLRYANYPKDYFMGMLPSSQYGSVAVLPS++S++ S + + + + S++ + +
Sbjct 241 TDMVQLRYANYPKDYFMGMLPSSQYGSVAVLPSLTSAAVSPTFIINKNNVSSSYTIVNNN 300
Query 301 gssssvrlsqtvsssQGIRLNSDLSALSIRATEYLQRWKEIVQFSSKDYSDQMAAQFGIK 360
++ + T Q +LN+DLSALS+RATEYLQRWKE+VQFSSKDYSDQMAAQFGIK
Sbjct 301 NNTLVASSTNTTGLEQYSQLNTDLSALSLRATEYLQRWKEVVQFSSKDYSDQMAAQFGIK 360
Query 361 APEYMGNHSHYIGGWSSVININEVVNTNLDADSSQASIAGKGISSNSGHTLTYDCGAEHQ 420
APEYMGNH+HYIGGWSSVININEVVNTNLD DSSQASIAGKG+SSNSGHT+TYDCGAEHQ
Sbjct 361 APEYMGNHAHYIGGWSSVININEVVNTNLDTDSSQASIAGKGVSSNSGHTITYDCGAEHQ 420
Query 421 IIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQSVPSLNLQNNPGRNVSGA 480
+IMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQ+VP+LNLQNNPGRNVSG+
Sbjct 421 VIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQAVPALNLQNNPGRNVSGS 480
Query 481 LGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGWNVLTSSGAWSYQSMKVRPQQLNS 540
LGYNLRYWQWKSNIDTVHAGFRAGAAYQSW APLDGW VLT+SG WSYQSMK+RPQQLNS
Sbjct 481 LGYNLRYWQWKSNIDTVHAGFRAGAAYQSWAAPLDGWQVLTASGTWSYQSMKIRPQQLNS 540
Query 541 IFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 579
IFVPQ+D+ANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY
Sbjct 541 IFVPQIDAANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 579
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 338 bits (868), Expect = 3e-109, Method: Compositional matrix adjust.
Identities = 242/629 (38%), Positives = 338/629 (54%), Gaps = 65/629 (10%)
Query 1 MSDFNPLNRARISTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVP 60
M+ + L+ + HRS FD+ K FTAKVGE+LP YW I++PG+KYR + ++FTRT P
Sbjct 1 MAFYTGLSNLQNHPHRSGFDIGRKNAFTAKVGELLPVYWDISMPGDKYRFNVEYFTRTQP 60
Query 61 VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDY--ItsaasstansstltsVPFVS 118
V T+AYTR++EY+DFYAVPLRL+ ++ P TQM D I + + + S
Sbjct 61 VATSAYTRLREYFDFYAVPLRLLWKSAPSVLTQMQDVNQIQALSLTQNLSLGTYLPSLTI 120
Query 119 QTLFNAFFQTANAGDQPNT----RDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITK 174
TL A +P T R+ G S KLL LGYG++I + ++
Sbjct 121 GTLGWAIRYLNGNTWEPETASYLRNAFGFSRADLSFKLLSYLGYGNLIET----PPSLGN 176
Query 175 KYLGVDSLSDADNPLVYQT----SQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDY 230
++ SL + D+ Y + VN P L YQKIY DFF QWEK +YNVDY
Sbjct 177 RWWST-SLKNTDDGANYTQQYIQNTIVNIFPLLTYQKIYQDFFRWPQWEKSNPSSYNVDY 235
Query 231 WSGAGNIGLVTD-------------MVQLRYANYPKDYFMGMLPSSQYGSVAVLPsisss 277
+SG+ + +V+D M L+Y N+ KD MG+LP+SQ+G VAVL ISSS
Sbjct 236 FSGS-SPSIVSDLPAASSAYWKSDTMFDLKYCNWNKDMLMGVLPNSQFGDVAVL-DISSS 293
Query 278 sdsrsLLYYEPsssataalqsaggssssvrlsqtvsssQ-----GIRLNSDLSA------ 326
DS +L +P S + S+ V L +S+ G +L+ DLS+
Sbjct 294 GDSDVVLGVDPHKSTLGIGSAITSKSAVVPLFALDASTSNPVSVGSKLHLDLSSIKSQFN 353
Query 327 -LSIRATEYLQRWKEIVQFSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVV 385
L++R E LQRWKEI Q DY +Q+ FG+K P+ + N YIGG S ++I+EVV
Sbjct 354 VLALRQAEALQRWKEISQSGDSDYREQILKHFGVKLPQALSNLCTYIGGISRNLDISEVV 413
Query 386 NTNLDADSSQASIAGKGISSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTV 445
N NL A+ A IAGKG+ + +G + TY EH +IM +YHAVP+LD+ LTGQ QL V
Sbjct 414 NNNLAAEEDTAVIAGKGVGTGNG-SFTYTTN-EHCVIMGIYHAVPLLDYTLTGQDGQLLV 471
Query 446 TAISDFPQPAFDQLGMQSVPSLNLQNN---PGRNVSGALGYNLRYWQWKSNIDTVHAGFR 502
T P P FD +G++ +P + N+ N+ A GYN RY+ WK+ +D ++ F
Sbjct 472 TDAESLPIPEFDNIGLEVLPMAQIFNSSLATAFNLFNA-GYNPRYFNWKTKLDVINGAFT 530
Query 503 AGAAYQSWVAP-----LDGWNVLTSS-------GAWSYQSMKVRPQQLNSIFVPQVDSAN 550
+SWV+P L GW +S +Y+ KV P L+ IF + DS
Sbjct 531 --TTLKSWVSPVSESLLSGWARFGASDSKTGTKAVLNYKFFKVNPSVLDPIFGVKADSTW 588
Query 551 CSVAFDQLLCNVNFQVYAVQNLDRNGLPY 579
+ DQLL N Y V+NL R+G+PY
Sbjct 589 DT---DQLLVNSYIGCYVVRNLSRDGVPY 614
> Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1
Length=607
Score = 338 bits (866), Expect = 5e-109, Method: Compositional matrix adjust.
Identities = 225/612 (37%), Positives = 315/612 (51%), Gaps = 60/612 (10%)
Query 15 HRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVPVNTAAYTRIKEYYD 74
HRS FD+ K FTAKVGE+LP YW I++PG+KY+ + ++FTRT PV T+AYTR++EY+D
Sbjct 9 HRSGFDIGRKNAFTAKVGELLPVYWDISMPGDKYKFNVEYFTRTQPVETSAYTRLREYFD 68
Query 75 FYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQTL--FNAFFQTANAG 132
FYAVPLRL+ ++ P TQM D I + + + F S TL F A N G
Sbjct 69 FYAVPLRLLWKSAPSVLTQMQD-INQLQALSFTQNLSLGSYFPSLTLSRFTAVLNRLNGG 127
Query 133 -----DQPNTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKKYLGVDSLSDADN 187
+ ++ G + KL LGYG++ +S S + ++
Sbjct 128 SNVPGNSSTFLNEFGFSRADLAFKLFSYLGYGNVWSSEFSS----SNRWWSTSLKGGGSY 183
Query 188 PLVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGAGNIGLVTD----- 242
Y VN P LAYQKIY DFF SQWE +YNVDY++G + LV+
Sbjct 184 TQQYVQDSYVNLFPLLAYQKIYQDFFRWSQWENSNPSSYNVDYFTGVSPL-LVSSLPEAS 242
Query 243 --------MVQLRYANYPKDYFMGMLPSSQYGSVAVLPsissssdsrsL----------- 283
M L+Y N+ KD MG+LP+SQ+G VAVL +S L
Sbjct 243 NAYWKSPTMFDLKYCNWNKDMLMGVLPNSQFGDVAVLDIDNSGKPDVVLGLGNANSTVGV 302
Query 284 LYYEPsssataalqsaggssssvrlsqtvsssQGIRLNSDLSALSIRATEYLQRWKEIVQ 343
Y S++A+ + SS++ + L S + L++R E LQRWKEI Q
Sbjct 303 ASYVSSNTASIPFFALKASSANTLPVGSTLRVDLASLKSQFTVLALRQAEALQRWKEISQ 362
Query 344 FSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNLDADSSQASIAGKGI 403
DY +Q+ FG+ P+ + N YIGG S ++I+EVVN NL A+ A IAGKG+
Sbjct 363 SGDSDYREQIRKHFGVNLPQALSNMCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGV 422
Query 404 SSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQS 463
+ +G + TY EH ++MC+YHAVP+LD+ +TGQ QL VT P P FD +GM++
Sbjct 423 GAGNG-SFTYTTN-EHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMET 480
Query 464 VPSLNLQNNPGRNVSGAL--GYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAP-----LDG 516
+P + N+P ++ GYN RY+ WK+ +D ++ F +SWV+P L G
Sbjct 481 LPMTQIFNSPKASIVNLFNAGYNPRYFNWKTKLDVINGAFT--TTLKSWVSPVTESLLSG 538
Query 517 WNVLTSSGA---------WSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVY 567
W S +Y+ KV P L+ IF DS S DQLL N Y
Sbjct 539 WFGFGYSEGDVNSQNKVVLNYKFFKVNPSVLDPIFGVAADSTWDS---DQLLVNSYIGCY 595
Query 568 AVQNLDRNGLPY 579
+NL R+G+PY
Sbjct 596 VARNLSRDGVPY 607
> Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1
Length=622
Score = 331 bits (849), Expect = 2e-106, Method: Compositional matrix adjust.
Identities = 229/629 (36%), Positives = 329/629 (52%), Gaps = 57/629 (9%)
Query 1 MSDFNPLNRARISTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVP 60
M+ F L + H++ FD+ SK LFTAKVGE++P YW AIP Y I +FTRT P
Sbjct 1 MAHFTGLKELQNHPHKAGFDVGSKNLFTAKVGELIPVYWDFAIPDCDYDIDLAYFTRTRP 60
Query 61 VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYItsaasstan-sstltsVPFVSQ 119
V TAAYTR++EY+DFYAVP L+ ++ A QM + + + + +P+ +
Sbjct 61 VQTAAYTRVREYFDFYAVPCDLLWKSFDSAVIQMGQTAPVQSKTLLDPLTVGNDIPWCTL 120
Query 120 -TLFNA-FFQTANAGDQPNTRDDAGLPIVYG------SCKLLDMLGYGSMIASNNPSKAA 171
L NA +F + ++ +G ++G KLL L YG+ + + + +
Sbjct 121 LDLSNAVYFSSGSSPLGSTVSVPSGFGNIFGYNRGDVDSKLLFYLNYGNFVNPSLSNVGS 180
Query 172 ITKKYLGVDSLSDADNP---LVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNV 228
+ ++ S S + P Y + VN P LAYQKIY DFF SQWE +YNV
Sbjct 181 PSNRWWNT-SFSSSKVPGYSQKYLNNNAVNIFPLLAYQKIYQDFFRWSQWENADPTSYNV 239
Query 229 DYWSGAGNI-------GLVT---------DMVQLRYANYPKDYFMGMLPSSQYGSVAVLP 272
DY++G+GN+ G VT +M LRY N+ KD F G+LP+SQ+G VAV+
Sbjct 240 DYYNGSGNLFGSSGLSGSVTASNSYWKRDNMFSLRYCNWNKDMFTGLLPNSQFGDVAVVN 299
Query 273 sissssdsrsLLYYEPsssataalqsaggssssvrlsqtvsssQGI--------RLN--- 321
S S + + + + T A + S+ S +S S + R+N
Sbjct 300 LGDSGSGTIPVGFLSDTEVFTQAFNATSMSTVSDTSPMGISGSTPVSARQSMVARINNAD 359
Query 322 -SDLSALSIRATEYLQRWKEIVQFSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVIN 380
+ S L++R E LQ+WKEI Q +Y DQ+ A FGI P M + + YIGG + ++
Sbjct 360 VASFSILALRQAEALQKWKEITQSVDTNYRDQIKAHFGINTPASMSHMAQYIGGVARNLD 419
Query 381 INEVVNTNLDADSSQASIAGKGISSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQA 440
I+EVVN NL D S+A I + + Y G+++ IIMC+YHA+P+LD+ +TGQ
Sbjct 420 ISEVVNNNLKDDGSEAVIY-GKGVGSGSGKMRYHTGSQYCIIMCIYHAMPLLDYAITGQD 478
Query 441 PQLTVTAISDFPQPAFDQLGMQSVPSLNLQNN---PGRNVSGALGYNLRYWQWKSNIDTV 497
PQL T++ D P P FD +GM++VP+ L N+ G V+ LGYN RYW WKS ID V
Sbjct 479 PQLLCTSVEDLPIPEFDNIGMEAVPATTLFNSVLFDGTAVNDFLGYNPRYWPWKSKIDRV 538
Query 498 HAGFRAGAAYQSWVAPLD-----GW--NVLTSSGAWSYQSMKVRPQQLNSIFVPQVDSAN 550
H F + WVAP+D W + S + S+ KV P L+SIF DS
Sbjct 539 HGAFT--TTLKDWVAPIDDDYLHNWFNSKDGKSASISWPFFKVNPNTLDSIFAVVADSI- 595
Query 551 CSVAFDQLLCNVNFQVYAVQNLDRNGLPY 579
DQLL N + V+ L ++G+PY
Sbjct 596 --WETDQLLINCDVSCKVVRPLSQDGMPY 622
> Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1
Length=602
Score = 330 bits (845), Expect = 4e-106, Method: Compositional matrix adjust.
Identities = 231/604 (38%), Positives = 320/604 (53%), Gaps = 57/604 (9%)
Query 16 RSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVPVNTAAYTRIKEYYDF 75
RS FD S FTAK GE+LP YW++ +PG K ++ FTRT+PVNTAAYTR+KEYYD+
Sbjct 16 RSGFDWSEHFSFTAKAGELLPVYWKMLLPGTKVNLNLSSFTRTMPVNTAAYTRVKEYYDW 75
Query 76 YAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQTLFNAFFQTANAGDQP 135
Y VPLRLI++++ QA QM D A S AN S +P+ + AN
Sbjct 76 YFVPLRLINKSIGQALVQMQDQPVQATSIVANKSVTLDLPWTNAATMFTLLNYANV-ILT 134
Query 136 NTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKKYLGVDSLSDADNPLVYQTSQ 195
N + G S KLL L YG+ +++PSK K + G+ S D + L+ +
Sbjct 135 NKYNLDGFSKAATSAKLLRYLRYGNCYYTSDPSKVGKNKNF-GLSSKDDFN--LLAAKNV 191
Query 196 TVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGAGNIGLVT----------DMVQ 245
+ N LP AYQKIY D+F QWE Y YN DY++G VT +++
Sbjct 192 SFNVLPLAAYQKIYCDWFRFEQWENACPYTYNFDYYNGGNVFAGVTANPENFWSNDNILS 251
Query 246 LRYANYPKDYFMGMLPSSQYGSVAVLPsissssdsrsLLYYEPsssataalqsaggssss 305
LRYANY KD FMG++PSSQ+GSVA + + SS + S P + + +++S
Sbjct 252 LRYANYNKDLFMGVMPSSQFGSVATVNVANFSSSNLS----SPLRNLSNVGMVTASNNAS 307
Query 306 vrlsqtvsssQ--------GI-------RLNSDLSALSIRATEYLQRWKEIVQFSSKDYS 350
+ SSQ GI L++ LS R E Q+WKE+ Q + + Y
Sbjct 308 SGSPLILRSSQDTSAGDGFGILTSNIFSTLSASFDILSFRIAEATQKWKEVTQCAKQGYK 367
Query 351 DQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNLDADSSQASIAGKGISSNSGHT 410
+Q+ A F +K E + +H YIGG SS + I+EV+NTNL +S+ A I GKG+ + G
Sbjct 368 EQLEAHFNVKLSEALSDHCRYIGGTSSGVTISEVLNTNL--ESAAADIKGKGVGGSFGSE 425
Query 411 LTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQSVPSLNLQ 470
T++ EH I+MC+YHA P+LD+ +GQ QL T +D P P FD +GM+++P L
Sbjct 426 -TFETN-EHGILMCIYHATPVLDYLRSGQDLQLLSTLATDIPIPEFDHIGMEALPIETLF 483
Query 471 NNPGRNVSG------ALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGWNVLTS-- 522
N + LGY+ RY +K+++D V F SWVAPL +T
Sbjct 484 NEQSTEATALINSIPVLGYSPRYIAYKTSVDWVSGAFE--TTLDSWVAPLTVNEQITKLL 541
Query 523 ----SG---AWSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRN 575
SG + +Y KV P+ L+ IFV + S DQ L NV+F V VQNLD N
Sbjct 542 FNPDSGSVYSMNYGFFKVTPRVLDPIFVQECTDTWDS---DQFLVNVSFNVKPVQNLDYN 598
Query 576 GLPY 579
G+PY
Sbjct 599 GMPY 602
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 317 bits (812), Expect = 6e-101, Method: Compositional matrix adjust.
Identities = 222/634 (35%), Positives = 332/634 (52%), Gaps = 71/634 (11%)
Query 1 MSDFNPLNRARISTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVP 60
MS FN ++ + RS FDLS++ FT+K GE+LP +W I PG+ ++I + FTRT P
Sbjct 1 MSLFN-MSAVKNHPRRSGFDLSNRVCFTSKAGELLPVFWDIVYPGDSFKIKTQLFTRTQP 59
Query 61 VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFV--- 117
+NTAAYTRI+EY DFY VPLRLI++ LP A TQM D A ++N T +P+V
Sbjct 60 LNTAAYTRIREYLDFYFVPLRLINKNLPTALTQMQDNPVQATGLSSNKVVTTDIPWVPVH 119
Query 118 ----SQTLFNAFFQTANAGDQPNTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAIT 173
S + F + + + G + S KLL L YG+ ++S P +
Sbjct 120 SSSGSYSSLTGFADVRGSVSSSDIENFLGFDSITQSAKLLMYLRYGNFLSSVVPDE---- 175
Query 174 KKYLGVDSLSDADNPLVYQTSQT-VNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWS 232
+K +G+ S D N T T V+ LP YQKIY DFF +QWEK++ Y YN D++S
Sbjct 176 QKSIGLSSSLDLRNSETVSTGYTSVHILPLATYQKIYADFFRFTQWEKNQPYTYNFDWYS 235
Query 233 GAGNIGLVT------------DMVQLRYANYPKDYFMGMLPSSQYGSVAVLPsissssds 280
G + +T ++ LRYAN+PKD FMG++P SQ G V+++ + S
Sbjct 236 GGNVLASLTTQALAKKYYSDDNLFTLRYANWPKDMFMGVMPDSQLGDVSIVDTSGSEGTF 295
Query 281 rsLLY----------------YEPsssataalqsaggssssvrlsqtvsssQGIRLNSDL 324
LY P++ ++ +Q+ S+S + G L S
Sbjct 296 PVGLYNFADGGSRAGLVAVSGTSPAAGSSLDMQTTSALSASTKYGVYAQQVAG--LGSSF 353
Query 325 SALSIRATEYLQRWKEIVQFSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEV 384
S L +R E +Q+++E+ QF+ +D Q+ A FG+ + + Y+GG SS I+++EV
Sbjct 354 SILQLRMAEAVQKYREVSQFADQDARGQIMAHFGVSLSPVLSDKCMYLGGSSSNIDLSEV 413
Query 385 VNTNLDADSSQASIAGKGISSNSGH-TLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQL 443
VNTN+ D+ A IAGKG+ + G + +D + IIM +YH VP+LD+ +TGQ L
Sbjct 414 VNTNITGDNI-AEIAGKGVGTGQGSFSGNFDT---YGIIMGIYHNVPLLDYVITGQPQNL 469
Query 444 TVTAISDFPQPAFDQLGMQSVPSLNLQNNPGRN-VSG------ALGYNLRYWQWKSNIDT 496
T +D P P +D +GMQ++ N+ SG +GY R++ K+ D
Sbjct 470 LYTNTADLPFPEYDSIGMQTIQFGRFVNSKAVGWTSGVDYRTQTMGYLPRFFDVKTRYDE 529
Query 497 VHAGFRAGAAYQSWVAPLDGWNV-------LTSSGA----WSYQSMKVRPQQLNSIFVPQ 545
V FR + ++WVAPLD N+ +TSSG +Y KV P+ L+SIF +
Sbjct 530 VLGAFR--STLKNWVAPLDPANLPQWLQTSVTSSGKLFLNLNYGFFKVNPRVLDSIFNVK 587
Query 546 VDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 579
DS ++ DQ L + + AV+N D +G+PY
Sbjct 588 CDS---TIDTDQFLTTLYMDIKAVRNFDYDGMPY 618
> Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1
Length=614
Score = 309 bits (791), Expect = 6e-98, Method: Compositional matrix adjust.
Identities = 217/619 (35%), Positives = 322/619 (52%), Gaps = 67/619 (11%)
Query 15 HRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVPVNTAAYTRIKEYYD 74
RS FDLS++ FT+K GE+LP +W I PG+ ++I + FTRT P+NTAAYTRI+EY D
Sbjct 9 RRSGFDLSNRVCFTSKAGELLPVFWDIVYPGDSFKIKTQLFTRTQPLNTAAYTRIREYLD 68
Query 75 FYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQTLFNAFFQTANAGDQ 134
FY VPLRLI++ LP A QM D A ++N T +P+V L + D
Sbjct 69 FYFVPLRLINKNLPTALMQMQDNPVQATGLSSNKVVTTDIPWVPTNLSGTYGSLTALADV 128
Query 135 PNT--------RDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKKYLGVDSLSDAD 186
N+ D G + S KLL L YG+ ++S K K LG+ D
Sbjct 129 KNSFPSSSTGIEDLLGFDAITQSAKLLMYLRYGNFLSSVVSDK----NKSLGLSGSLDLR 184
Query 187 NPLVYQTSQT-VNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGAGNIGLVT---- 241
N T T ++ LP AYQK Y DFF +QWEK++ Y YN D++SG + +T
Sbjct 185 NSETASTGYTSMHILPLAAYQKAYADFFRFTQWEKNQPYTYNFDWYSGGNVLASLTTLDL 244
Query 242 --------DMVQLRYANYPKDYFMGMLPSSQYGSVAV--------------LPsissssd 279
++ LRYAN+PKD FMG++P SQ G V++ L +
Sbjct 245 AKKYYSDDNLFTLRYANWPKDMFMGVMPDSQLGDVSIVDASGSEGTFPVGLLDVNDGTLR 304
Query 280 srsLLYYEPsssataalqsaggssssvrlsqtvsssQGIRLNSDLSALSIRATEYLQRWK 339
+ L + + ++L+ S+ S + V + + L S S L +R E +Q+++
Sbjct 305 AGLLARSGSAPAEKSSLEMQTSSALSANTTYGVYAQRAAGLASSFSILQLRMAEAVQKYR 364
Query 340 EIVQFSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNLDADSSQASIA 399
E+ QF+ +D Q+ A FG+ + + Y+GG SS I+++EVVNTN+ D+ A IA
Sbjct 365 EVSQFADQDARGQIMAHFGVSLSPVLSDKCVYLGGSSSNIDLSEVVNTNITGDNV-AEIA 423
Query 400 GKGISSNSGH-TLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQ 458
GKG+ + G + +D E+ II+ +YH VP+LD+ +TGQ L T +D P P FD
Sbjct 424 GKGVGTGQGSFSGQFD---EYGIIIGIYHNVPLLDYVITGQPQNLLYTNTADLPFPEFDS 480
Query 459 LGMQSVPSLNLQNNPGRN-VSG------ALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWV 511
+GMQ++ N+ + SG +GY R++ K+ D V FR + ++WV
Sbjct 481 IGMQTIQFGRFVNSKSVSWTSGVDYRVQTMGYLPRFFDVKTRYDEVLGAFR--STLKNWV 538
Query 512 APLD-----GW--NVLTSSGAWS----YQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLC 560
APLD W + +TSSG + Y KV P+ L+SIF + DS ++ DQ L
Sbjct 539 APLDPSYVSKWLQSSVTSSGKLALNLNYGFFKVNPRVLDSIFNVKCDS---TIDTDQFLT 595
Query 561 NVNFQVYAVQNLDRNGLPY 579
+ + AV+N D +G+PY
Sbjct 596 ALYMDIKAVRNFDYDGMPY 614
> Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1
Length=622
Score = 304 bits (778), Expect = 5e-96, Method: Compositional matrix adjust.
Identities = 219/625 (35%), Positives = 315/625 (50%), Gaps = 76/625 (12%)
Query 15 HRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVPVNTAAYTRIKEYYD 74
RS FDL+ + FT+K GE+LP YW+ IPG+K+ IS++WFTRT PV+TAAYTR+KEY D
Sbjct 14 RRSGFDLTKRICFTSKAGELLPVYWKPTIPGDKWNISTNWFTRTQPVDTAAYTRVKEYVD 73
Query 75 FYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQTLFNAFFQTANAGDQ 134
++ VPL LI + + A TQM D SA S+ N + T +P+ + + N
Sbjct 74 WFFVPLNLIQKGIESAITQMVDNPVSAMSAIENRAITTDMPYTTLLSLSRALYMLNGKSY 133
Query 135 PNTRDDAGLPIVYG------SCKLLDMLGYGSMIASNNPSKAAITKKYLGVDSLSDADNP 188
N+ AG ++G S KLL ML YG+ I NP + + G ++ A
Sbjct 134 VNSH--AGKLNMFGFSRADLSAKLLQMLKYGNFI---NPEHSGLDTPMFGYSTVKLAQ-- 186
Query 189 LVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGAGNIGLVTDM----- 243
Y +Q VN LP YQKIY D+F QWEK Y YN DY++G T M
Sbjct 187 FSYLWNQAVNVLPIFCYQKIYSDYFRFQQWEKPVPYTYNADYYAGGDIFPNPTTMPDAYW 246
Query 244 -----VQLRYANYPKDYFMGMLPSSQYG--SVAVLPsissssdsrsLLYYEPsssataal 296
LRY N+ +D + G LP+ Q+G SV + S + + + +
Sbjct 247 NNYTPFDLRYCNWNRDLYTGFLPNQQFGNVSVVDMTVSSDEIMAAPVTFGATGNRVAVTK 306
Query 297 qsaggssssvrlsqtvsssQGI--------------RLNSDLSALSIRATEYLQRWKEIV 342
+ S+SS + + S G L S L++R E+LQ+WKEI
Sbjct 307 AALASSTSSTGIGTVSTPSGGTVPVGTSLYAQVQQRDLTGAFSILALRQAEFLQKWKEIA 366
Query 343 QFSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNL------DADSSQA 396
+DY Q+ FG+K P + S YIGG + ++I+EVVN NL DA A
Sbjct 367 LSGDQDYRSQIEKHFGVKLPAELSYMSQYIGGQFAQMDISEVVNQNLTDQAGSDAAQYPA 426
Query 397 SIAGKGISSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAF 456
IAGKG++S G+ + Y +H IIM +YHAVP+LD+ TGQ L +T+ ++ P F
Sbjct 427 LIAGKGVNSGDGN-VNY-TARQHGIIMGIYHAVPLLDYERTGQDQDLLITSAEEWAIPEF 484
Query 457 DQLGMQSVPSLNLQNNPGRNVSG-----------ALGYNLRYWQWKSNIDTVHAGFRAGA 505
D +GMQ++P L N+ VSG +GY RY WK++ID + FR+
Sbjct 485 DAIGMQTLPLGTLFNS--NKVSGDSDFRLHGAAYPIGYVPRYVNWKTDIDEIFGAFRSSE 542
Query 506 AYQSWVAPLDG-----W--NVLTSSGA----WSYQSMKVRPQQLNSIFVPQVDSANCSVA 554
++WVAP+D W NV ++ A ++Y KV P L+ IF + DS ++
Sbjct 543 --KTWVAPIDADFITNWVKNVADNASAVQSLFNYNWFKVNPAILDDIFAVKADS---TMD 597
Query 555 FDQLLCNVNFQVYAVQNLDRNGLPY 579
D + + V+NLD +G+PY
Sbjct 598 TDTFKTRMMMSIKCVRNLDYSGMPY 622
> Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1
Length=584
Score = 254 bits (649), Expect = 1e-77, Method: Compositional matrix adjust.
Identities = 187/609 (31%), Positives = 296/609 (49%), Gaps = 55/609 (9%)
Query 1 MSDFNPLNRARISTHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVP 60
M FN L+ + RS FDLSSK F+AKVGE+LP W + +PG+K+ + FTRT P
Sbjct 1 MGLFN-LSSVKNRPRRSGFDLSSKVAFSAKVGELLPIKWTLTMPGDKFSLKEQHFTRTQP 59
Query 61 VNTAAYTRIKEYYDFYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQT 120
VNT+AYTR++EYYD++ PL L+ R P+ Q+ + A+S + +++P S
Sbjct 60 VNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQIQQNVQHASSFDGSVLLGSNMPCFSAD 119
Query 121 LFNAFFQTANAGDQPNTRDD-AGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKKYLGV 179
Q + + D ++ + G + KL+ L YG++ S + + Y
Sbjct 120 ------QISQSLDMMKSKLNYFGFNRADLAYKLIQYLRYGNVRTGVGTSGS---RNYGTS 170
Query 180 DSLSDADNPLVYQTSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGAGNIGL 239
+ D+ + ++ P LAY+K D+F +QW+ Y +N+DY+ G G +
Sbjct 171 VDVKDSSYNQNRAFNHALSVFPILAYKKFCQDYFRLTQWQVSAPYLWNIDYYDGKGATTI 230
Query 240 V-------------TDMVQLRYANYPKDYFMGMLPSSQYGSVAVLPsissssdsrsLLYY 286
+ L Y N+ KD F G LP +QYG +V+ ++ +
Sbjct 231 LPADLSKSVTYFEHDTFFDLEYCNWNKDMFFGSLPDAQYGDTSVVDISYGTTGA------ 284
Query 287 EPsssataalqsaggssssvrlsqtvsssQGIRLNSDLSALSIRATEYLQRWKEIVQFSS 346
P +A S++ + + N L L++R E LQR++EI +
Sbjct 285 -PVITAQNLQSPVNSSTAIGTSDKFSTQLIEAGTNLTLDVLALRRGEALQRFREISLCTP 343
Query 347 KDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSVININEVVNTNLDADSSQASIAGKGISSN 406
+Y Q+ A FG+ + S YIGG +S ++I+EVVNTN+ +S++A IAGKGI +
Sbjct 344 LNYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNTNI-TESNEALIAGKGIGTG 402
Query 407 SGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTGQAPQLTVTAISDFPQPAFDQLGMQSVPS 466
G+ Y + ++MC+YH+VP+LD+ ++ PQL + + FP P D +G++ +
Sbjct 403 QGNEEFY--AKDWGVLMCIYHSVPLLDYVISAPDPQLFASMNTSFPVPELDAIGLEPITV 460
Query 467 LNLQNNPGRNVSGA---------LGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPL--D 515
NNP S +GY RY+ WK++ID V F + WVAP+ +
Sbjct 461 AYYSNNPIELPSTGGITDAPTTTVGYLPRYYAWKTSIDYVLGAFT--TTEKEWVAPITPE 518
Query 516 GW-NVL----TSSGAWSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQ 570
W N+L T +Y KV P L+ IF DS + D L N F + +
Sbjct 519 LWSNMLKPLGTKGTGINYNFFKVNPSILDPIFAVNADSYWDT---DTFLINAAFDIRVAR 575
Query 571 NLDRNGLPY 579
NLD +G+PY
Sbjct 576 NLDYDGMPY 584
> Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1
Length=618
Score = 169 bits (427), Expect = 6e-47, Method: Compositional matrix adjust.
Identities = 100/268 (37%), Positives = 146/268 (54%), Gaps = 38/268 (14%)
Query 13 STHRSSFDLSSKKLFTAKVGEILPCYWQIAIPGNKYRISSDWFTRTVPVNTAAYTRIKEY 72
S RS FDLS+K FTAKVGE+LP YW+ +PG+K+ IS +WFTRT PV+T+A+TRI+EY
Sbjct 13 SPRRSGFDLSNKCAFTAKVGELLPVYWKFCLPGDKFNISQEWFTRTQPVDTSAFTRIREY 72
Query 73 YDFYAVPLRLISRALPQAFTQMTDYItsaasstansstltsVPFVSQTLFNAFFQTANAG 132
Y+++ VPL L+ R +A M + AAS +++ S ++P+V + N +
Sbjct 73 YEWFFVPLHLLYRNSNEAIMSMENQPNYAASGSSSISFNRNLPWVDLSTINVAIGNVQSS 132
Query 133 DQPNTRDDAGLPIVYGSCKLLDMLGYGSMIASNNPSKAAITKKYLGVDSLSDADNPLVYQ 192
P ++ G+ G KL+ LGYG +P Y
Sbjct 133 TSP--KNFFGVSRSEGFKKLVSYLGYGET-------------------------SPEKYV 165
Query 193 TSQTVNALPFLAYQKIYYDFFSNSQWEKHKAYAYNVDYWSGAGNIGLVT----------- 241
+ +A P AYQKIY D++ +SQWEK K + YN D+W+G + +
Sbjct 166 DNLRCSAFPLYAYQKIYQDYYRHSQWEKSKPWTYNCDFWNGEDSTPITATVELFSQSPND 225
Query 242 DMVQLRYANYPKDYFMGMLPSSQYGSVA 269
+ +LRYAN+ KD +MG LP+SQ+G VA
Sbjct 226 SVFELRYANWNKDLWMGSLPNSQFGDVA 253
Score = 162 bits (409), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 107/274 (39%), Positives = 147/274 (54%), Gaps = 20/274 (7%)
Query 319 RLNSDLSALSIRATEYLQRWKEIVQFSSKDYSDQMAAQFGIKAPEYMGNHSHYIGGWSSV 378
+L++ S L +RA E LQ+WKEI Q + ++Y+ Q+ A FG+ + S I G+
Sbjct 352 QLDAQFSVLQLRAAEALQKWKEIAQANGQNYAAQVKAHFGVSTNPMQSHRSTRICGFDGS 411
Query 379 ININEVVNTNLDADSSQASIAGKGISSNSGHTLTYDCGAEHQIIMCVYHAVPMLDWNLTG 438
I+I+ V NTNL AD +A I GKG+ + + EH +IMC+YHA P+LD+ TG
Sbjct 412 IDISAVENTNLTAD--EAIIRGKGLGGQRINDPSDFTCNEHGVIMCIYHATPLLDYVPTG 469
Query 439 QAPQLTVTAISD-FPQPAFDQLGMQSVPSLNLQNNP--GRNVSGA-LGYNLRYWQWKSNI 494
QL T + +P P FD LGM+S+P L+L N+ G V+ + GY RY WK++I
Sbjct 470 PDLQLMSTVKGESWPVPEFDSLGMESLPMLSLVNSKAIGDIVARSYAGYVPRYISWKTSI 529
Query 495 DTVHAGFRAGAAYQSWVAPLDG-------WNVLTSSGA--WSYQSMKVRPQQLNSIFVPQ 545
D V F +SW AP+D V+ G+ SY KV P LN IF
Sbjct 530 DVVRGAFT--DTLKSWTAPVDSDYMHVFFGEVIPQEGSPILSYTWFKVNPSVLNPIFAVS 587
Query 546 VDSANCSVAFDQLLCNVNFQVYAVQNLDRNGLPY 579
VD S DQLLCN F V +NL +G+PY
Sbjct 588 VDG---SWNTDQLLCNCQFDVKVARNLSYDGMPY 618
Lambda K H a alpha
0.319 0.133 0.409 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 53365796