bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-14_CDS_annotation_glimmer3.pl_2_2
Length=410
Score E
Sequences producing significant alignments: (Bits) Value
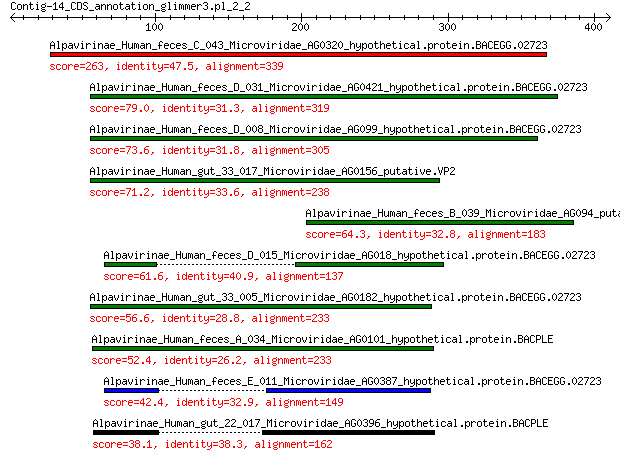
Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.... 263 4e-85
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 79.0 8e-18
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 73.6 5e-16
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 71.2 2e-15
Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2 64.3 4e-13
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 61.6 3e-12
Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.p... 56.6 1e-10
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 52.4 2e-09
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 42.4 5e-06
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 38.1 9e-05
> Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.protein.BACEGG.02723
Length=396
Score = 263 bits (673), Expect = 4e-85, Method: Compositional matrix adjust.
Identities = 161/346 (47%), Positives = 217/346 (63%), Gaps = 11/346 (3%)
Query 28 KMNKKSIKYNKWalqeqqrfqseqaqlGRDWSEEMMSKANQWNLEQWNRENEYNLPVNRK 87
KMN+++ KYNKWALQ+Q F +QAQLGRDWSEEMMS+ N+WNL+QWNREN YN P ++
Sbjct 29 KMNRRAEKYNKWALQQQMAFNEQQAQLGRDWSEEMMSQQNEWNLQQWNRENAYNTPAAQR 88
Query 88 ARLLAAQINPALAMQggssvgqaasspssasapspaspsgasaLPLNLQRPDYGTGFAQL 147
+RL AA +N ALAMQG S+G A S +A+ + + RPD F+ L
Sbjct 89 SRLEAAGLNAALAMQGQGSIGMAGSGQPAAAPAGSPQAATGGSSAPQYSRPD----FSLL 144
Query 148 SSAVNSYFENKQRDVITEGYGLDNALKATYGDRAYRLSLGKTEAEIDNIRASTAKSYADS 207
S AV+S+F+NK + G GLDN LKA YGD ++S+GK AEI N+R+ +A++YA++
Sbjct 145 SQAVDSFFKNKLLSEQSTGQGLDNLLKARYGDELAQISIGKGSAEISNLRSQSARNYAET 204
Query 208 ALVNLQAKEKEILNKYLDAGQQLSLFLKIGELATMKTQRELLSAQTRKAIAEEIEVSARA 267
A+ +L A + LNKYLD GQQLSL K+ E +++ EL A+ R IA EI+ A A
Sbjct 205 AVASLTADAQRTLNKYLDMGQQLSLITKMAEYSSITAGTELTKAKYRTEIANEIKTLAEA 264
Query 268 RGLKISNYIAEQTAERLIIATNEENRYRGIHARSSAAWTPVRDF-------YKTSQMSAD 320
G KISN IA TA+ LI A N+EN YR A + P R + Y+ + D
Sbjct 265 NGQKISNEIARSTAQSLIDAMNKENEYRSYDAALGYDYLPRRHYLKNKGLGYEIDLLEGD 324
Query 321 LKASEVANIMAEFERYTKDTPGNRWIQKNIVPVSSALGPLLDAAAM 366
L V +AEFE YT+DTPGNRW++KN+ P++ +G LL A +
Sbjct 325 LGLQRVERALAEFEEYTRDTPGNRWLRKNVDPITGIVGTLLGATTL 370
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 79.0 bits (193), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 100/354 (28%), Positives = 158/354 (45%), Gaps = 48/354 (14%)
Query 56 RDWSEEMMSKANQWNLEQWNRENEYNLPVNRKARLLAAQINPALAMQggssvgqaassps 115
RD+ + QWNL+QWNREN YN P ++AR+ AA NP M G + + S
Sbjct 57 RDFQTSEREASQQWNLDQWNRENAYNDPSAQRARMEAAGFNP-YNMNIDPGSGSTSGAQS 115
Query 116 sasapspaspsgasaLPL------NLQRPDYGTGFAQLSSAVNSYFENKQRDVITEGYGL 169
S + S A+ S +LP + Q + +G AQ+ +AV S + + +T YG
Sbjct 116 SPGSGSSATASHTPSLPAYTGYAADFQ--NVASGIAQIGNAVASGIDAR----LTSAYG- 168
Query 170 DNALKATYGDR----------AYRLS------------LGKTEAEIDNIRASTAKSYADS 207
D+ +KA + Y+L L K E + + + A
Sbjct 169 DDLMKADIMSKIGGNSEWLTDVYKLGRQNEAPNLLGIDLRKKRLENLSTETNIKVALAQG 228
Query 208 ALVNLQAKEKEILNKYLDAGQQLSLFLKIGELATMKTQRELLSAQTRKAIAEEIEVSARA 267
AL+ LQA+ + I+NK++ A QQ FLK +L AQ + I ++ + A+A
Sbjct 229 ALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSEAQVKTQIKQQALLEAQA 288
Query 268 RGLKISNYIAEQTAERLIIATNEENR-----YRGIHARSSAAWTPVRDFYKTSQMSADLK 322
G K++N +AE+ A+ A E R Y G + + AW K +Q+ +
Sbjct 289 AGQKLNNRLAERLADYQFKAMAAEYRANAAYYNGFY---NDAWQA--GMSKAAQVRYESN 343
Query 323 ASEVANIMAE-FERYTKDTPGNRWIQKNIVP-VSSALGPLLDAAAMFTVAGKLG 374
A+ +A M+E F+ K + N I NI + LG + F++A K+G
Sbjct 344 AARIAAQMSEIFKDREKASWKNNPIYYNISELLKGLLGSVGSVVGPFSIASKIG 397
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 73.6 bits (179), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 97/347 (28%), Positives = 151/347 (44%), Gaps = 55/347 (16%)
Query 56 RDWSEEMMSKANQWNLEQWNRENEYNLPVNRKARLLAAQINPALAMQggssvgqaassps 115
RD+ + QWNL+QWNREN YN P ++AR+ AA NP M + + + S
Sbjct 57 RDFQTSEREASQQWNLDQWNRENAYNDPSAQRARMEAAGFNP-YNMNIDAGSASTSGAQS 115
Query 116 sasapspaspsgasaLPL------NLQRPDYGTGFAQLSSAVNSYFENKQRDVITEGYGL 169
S + S A+ S +LP + Q + +G AQ+ +AV+S + + +T YG
Sbjct 116 SPGSGSQATASHTPSLPAYTGYAADFQ--NVASGIAQIGNAVSSGIDAR----LTSAYG- 168
Query 170 DNALKATYGDR----------AYRLS------------LGKTEAEIDNIRASTAKSYADS 207
D+ +KA + Y+L L K E + + + A
Sbjct 169 DDLMKADIMSKIGGNSEWLTDVYKLGRQNEAPNLLGIDLRKKRLENLSTETNIKVALAQG 228
Query 208 ALVNLQAKEKEILNKYLDAGQQLSLFLKIGELATMKTQRELLSAQTRKAIAEEIEVSARA 267
AL+ LQA+ + I+NK++ A QQ FLK +L AQ + I ++ + A+
Sbjct 229 ALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSEAQVKTQIKQQALLEAQT 288
Query 268 RGLKISNYIAEQTAERLIIATNEENR-----YRGIHARSSAAWTPVRDFYKTSQMSADLK 322
G K++N +AE+ A+ A E R Y G + + AW K +Q +
Sbjct 289 VGQKLNNRLAERLADYQFKAMAAEYRANAAYYNGFY---NDAWQA--GMSKAAQARYESN 343
Query 323 ASEVANIMAE-FERYTKDTPGNRWIQKNI--------VPVSSALGPL 360
A+ +A M+E F+ K + N I NI V S GPL
Sbjct 344 AARIAADMSEIFKGREKSSWKNNPIYYNIGELLKGILGSVGSVAGPL 390
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 71.2 bits (173), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 80/263 (30%), Positives = 117/263 (44%), Gaps = 48/263 (18%)
Query 56 RDWSEEMMSKANQWNLEQWNRENEYNLPVNRKARLLAAQINPALAMQggssvgqaassps 115
RD+ E M WN+EN YN ++ RL A +NP L M GGSS ++
Sbjct 54 RDFQENM-----------WNKENTYNTASAQRQRLEEAGLNPYLMMNGGSSGVSQSAGTG 102
Query 116 sasapspaspsgasaLPLNLQRPDYGTGFAQLSSAVNSYFENKQRDV-ITEGYGLDNA-- 172
++++ S + +P + F+ + A+ S F+++ R +++ G N
Sbjct 103 ASASSSGTAVF----------QP-FQADFSGIQQAIGSVFQSQVRQAQVSQMQGQRNLAD 151
Query 173 ----------------------LKATYGDRAYRLSLGKTEAEIDNIRASTAKSYADSALV 210
LKAT RA RL K E+DN+ + A
Sbjct 152 AQAMQALSQVDWSKMTKETREYLKATGLARA-RLGYSKEMQELDNMAFAGRLLQAQGTSQ 210
Query 211 NLQAKEKEILNKYLDAGQQLSLFLKIGELATMKTQRELLSAQTRKAIAEEIEVSARARGL 270
L+A K +LN+YLD QQ L +K +Q L Q +K IA+EI AR +G
Sbjct 211 LLEADAKTVLNRYLDQQQQADLNVKASVYYNQMSQGHLNYNQAKKVIADEILTYARIKGQ 270
Query 271 KISNYIAEQTAERLIIATNEENR 293
K+SN +AE TA+ LI ATN NR
Sbjct 271 KLSNKVAEATADSLIRATNAANR 293
> Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2
Length=380
Score = 64.3 bits (155), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 60/192 (31%), Positives = 85/192 (44%), Gaps = 30/192 (16%)
Query 203 SYADSALVNLQAKEKEILNKYLDAGQQLSLFLKIGELATMKTQRELLSAQTRKAIAEEIE 262
S A A + L + + I+NKY+D QQ LF+K LA +++Q L Q + I I
Sbjct 200 SAAQEAQILLNSDAQRIMNKYMDQNQQADLFIKAQTLANLQSQGALTEKQIQTEIQRAIL 259
Query 263 VSARARGLKISNYIAEQTAERLIIATNEENR---------YRGIHARSSAAWTPVRDFYK 313
SA A G KI N +A +TA+ LI A N N Y+ + R YK
Sbjct 260 ASAEASGKKIDNRVASETADSLIKAANASNELQYRDSTYDYKNVKLRKHTE-------YK 312
Query 314 TSQMSADLKASEVANIMAEFERYTKDTPGNRWIQKNIVPVSSALGPLLDAAAMFTVAGKL 373
TS A+ KA+E +A + T + W V+ +G + A F A +
Sbjct 313 TSM--ANQKAAEYGADLARKQGRT-----HYW-----ESVARGIGSIAAGAGNFIGAFRP 360
Query 374 GKSVNIFKPKYG 385
G NIF+ YG
Sbjct 361 G--ANIFRNDYG 370
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 61.6 bits (148), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 38/101 (38%), Positives = 56/101 (55%), Gaps = 7/101 (7%)
Query 196 IRASTAKSYADSALVNLQAKEKEILNKYLDAGQQLSLFLKIGELATMKTQRELLSAQTRK 255
+RA+ A S D A+ K ILNKYLD QQ L +K + + +L + R+
Sbjct 230 LRANVANSLLD-------AESKTILNKYLDQQQQADLNVKAAHYEELINRGQLHVVEARE 282
Query 256 AIAEEIEVSARARGLKISNYIAEQTAERLIIATNEENRYRG 296
++ E+ ARARGL ISN++A ++A+ L+ A N N Y G
Sbjct 283 LLSREVLNYARARGLNISNWVAAKSAKGLVYANNAANYYEG 323
Score = 32.3 bits (72), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 24/40 (60%), Gaps = 4/40 (10%)
Query 65 KANQWNLEQWNRENEYNLPVNRKARL----LAAQINPALA 100
KA + L+ WN+EN YN P ++ARL A +NPA A
Sbjct 79 KARAFQLDMWNKENAYNTPAAQRARLEEGGYNAYMNPADA 118
> Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.protein.BACEGG.02723
Length=354
Score = 56.6 bits (135), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 67/235 (29%), Positives = 107/235 (46%), Gaps = 27/235 (11%)
Query 56 RDWSEEMMSKANQWNLEQWNRENEYNLPVNRKARLLAAQINPALAMQggssvgqaassps 115
RD+++ M N+WN + N++ARL AA +NP L M GGS+ +++S S
Sbjct 52 RDFAKSMFDATNEWNSAK-----------NQRARLEAAGLNPYLMMNGGSAGTASSTSAS 100
Query 116 sasapspaspsgasaLPLNL--QRPDYGTGFAQLSSAVNSYFENKQRDVITEGYGLDNAL 173
+ S S + + P N+ Y + +S A R TE LD
Sbjct 101 TVSGASGSGGTPYQYTPTNMIGDVASYASAMKSMSDA---------RKTNTESDLLD--- 148
Query 174 KATYGDRAYRLSLGKTEAEIDNIRASTAKSYADSALVNLQAKEKEILNKYLDAGQQLSLF 233
YG Y +GKT A+ + + A A + L+ +E+LN YL +++ L
Sbjct 149 --KYGVPTYESQIGKTMADTYFTQRQADVAIAQKANLLLRNDAQEVLNMYLPEEKRIQLQ 206
Query 234 LKIGELATMKTQRELLSAQTRKAIAEEIEVSARARGLKISNYIAEQTAERLIIAT 288
+ + M + + Q + IA +E+ AR +G ISN IA+ TA+ +I AT
Sbjct 207 MNGAQYWNMIREGVISEEQAKNLIASRLEIEARTQGQHISNKIAKSTADSIIDAT 261
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 52.4 bits (124), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 61/235 (26%), Positives = 103/235 (44%), Gaps = 16/235 (7%)
Query 57 DWSEEMMSKANQWNLEQWNRENEYNLPVNRKARLLAAQINPALAMQggssvgqaasspss 116
+W+ K+ + ++ NE+N +++ARL A +NP L M GGS+ ++S ++
Sbjct 42 EWASSESQKSRDFAKSMFDASNEWNSAKSQRARLEEAGLNPYLMMNGGSAGTAQSTSATA 101
Query 117 asapspaspsgasaLPLNL--QRPDYGTGFAQLSSAVNSYFENKQRDVITEGYGLDNALK 174
+S S + P N+ Y LS A S G + L
Sbjct 102 SSGSSGSGGMPYQYTPTNVIGDVASYAGAMKSLSDARKS--------------GTEADLL 147
Query 175 ATYGDRAYRLSLGKTEAEIDNIRASTAKSYADSALVNLQAKEKEILNKYLDAGQQLSLFL 234
YGD Y + TEA+ + + + A A + L A+ ++++N YL +Q+ L
Sbjct 148 GRYGDSDYSSRIANTEADTYFKQRQSDVATAQKANLLLSAEAQQVMNMYLPQEKQIQLST 207
Query 235 KIGELATMKTQRELLSAQTRKAIAEEIEVSARARGLKISNYIAEQTAERLIIATN 289
+ M + Q + +A +E+ AR G ISN +A TA+ +I ATN
Sbjct 208 LGAQYWNMIRDGSIKEEQAKNLLATRLEIEARTAGQHISNKVARSTADSIIDATN 262
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 42.4 bits (98), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 33/112 (29%), Positives = 51/112 (46%), Gaps = 0/112 (0%)
Query 176 TYGDRAYRLSLGKTEAEIDNIRASTAKSYADSALVNLQAKEKEILNKYLDAGQQLSLFLK 235
T G +LS + ++N++ A + L + K I+NKYLD Q L L L
Sbjct 219 TTGMARAQLSFNTDQQNLENMKWVNKIQRAQRTDILLSNQAKGIINKYLDTSQSLQLKLM 278
Query 236 IGELATMKTQRELLSAQTRKAIAEEIEVSARARGLKISNYIAEQTAERLIIA 287
+ L Q + + +++ A G KISN IA +TA++LI A
Sbjct 279 ANQSFQAFASGRLSLQQCKTEVTKQLMNMAETEGKKISNKIASETADQLIGA 330
Score = 33.9 bits (76), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 65 KANQWNLEQWNRENEYNLPVNRKARLLAAQINPALAM 101
KA Q+ E WN+ N++N P N + RL A NP L +
Sbjct 93 KARQYQSEMWNKTNDWNSPKNVRKRLQEAGYNPYLGL 129
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 38.1 bits (87), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 46/121 (38%), Positives = 63/121 (52%), Gaps = 4/121 (3%)
Query 173 LKATYGDRAYRLSLGKTEAEIDNIRASTAKSYADSALVNLQAKEKEILNKYLDAGQQLSL 232
LK+T RA +L K + E DN+ + A + + L + K ILNKYLD QQL L
Sbjct 174 LKSTGLARA-QLGYAKEQQEADNMAMTGLVLRAQRSGMLLDNEAKGILNKYLDQHQQLDL 232
Query 233 FLKIGELATMKTQRELLSAQTRKAIAEEIEVSARARGLKISNYIAEQTAERLI---IATN 289
+K + + A+ +KA+AEE +AR RG ISN +A + AE I IA N
Sbjct 233 SVKAADYYQRMAAGYVSYAEAKKALAEEALAAARTRGQNISNEVASRIAESQIAANIAAN 292
Query 290 E 290
E
Sbjct 293 E 293
Score = 33.5 bits (75), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 58 WSEEMMSKANQWNLEQWNRENEYNLPVNRKARLLAAQINPALAM 101
++E M + W WN+EN YN ++ RL A +NP L M
Sbjct 45 FNERMAIQQRNWQENMWNKENAYNTASAQRQRLEEAGLNPYLMM 88
Lambda K H a alpha
0.312 0.127 0.361 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 36142824