bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-12_CDS_annotation_glimmer3.pl_2_4
Length=72
Score E
Sequences producing significant alignments: (Bits) Value
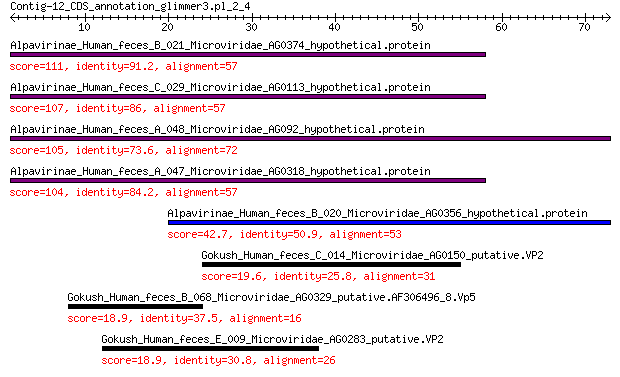
Alpavirinae_Human_feces_B_021_Microviridae_AG0374_hypothetical.... 111 5e-35
Alpavirinae_Human_feces_C_029_Microviridae_AG0113_hypothetical.... 107 1e-33
Alpavirinae_Human_feces_A_048_Microviridae_AG092_hypothetical.p... 105 2e-32
Alpavirinae_Human_feces_A_047_Microviridae_AG0318_hypothetical.... 104 2e-32
Alpavirinae_Human_feces_B_020_Microviridae_AG0356_hypothetical.... 42.7 7e-09
Gokush_Human_feces_C_014_Microviridae_AG0150_putative.VP2 19.6 4.6
Gokush_Human_feces_B_068_Microviridae_AG0329_putative.AF306496_... 18.9 7.0
Gokush_Human_feces_E_009_Microviridae_AG0283_putative.VP2 18.9 8.7
> Alpavirinae_Human_feces_B_021_Microviridae_AG0374_hypothetical.protein
Length=66
Score = 111 bits (277), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 52/57 (91%), Positives = 56/57 (98%), Gaps = 0/57 (0%)
Query 1 MAATKFTAIYVNNDGKLIEKEIPGMNTYKIAEKFAIMLNDTEETKVVCVIESWKLYP 57
MAATKFTAIYVNN+GK+IE+EIPGMNTYKIAEKFAIMLND EETK+VCVIESWKLYP
Sbjct 1 MAATKFTAIYVNNEGKIIEREIPGMNTYKIAEKFAIMLNDPEETKLVCVIESWKLYP 57
> Alpavirinae_Human_feces_C_029_Microviridae_AG0113_hypothetical.protein
Length=73
Score = 107 bits (268), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 49/57 (86%), Positives = 56/57 (98%), Gaps = 0/57 (0%)
Query 1 MAATKFTAIYVNNDGKLIEKEIPGMNTYKIAEKFAIMLNDTEETKVVCVIESWKLYP 57
MAATKFTAIYVNN+GK+IE+EIPGMNTYKIAEKFA+MLND +ETK+VCV+ESWKLYP
Sbjct 1 MAATKFTAIYVNNEGKIIEREIPGMNTYKIAEKFAMMLNDPKETKLVCVVESWKLYP 57
> Alpavirinae_Human_feces_A_048_Microviridae_AG092_hypothetical.protein
Length=73
Score = 105 bits (261), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 53/72 (74%), Positives = 65/72 (90%), Gaps = 0/72 (0%)
Query 1 MAATKFTAIYVNNDGKLIEKEIPGMNTYKIAEKFAIMLNDTEETKVVCVIESWKLYPnen 60
MAATKFTAIY+NN+GK+I++EIPGMN+YKIAEKFA+MLND EETK+VCVIE+WK+YPN
Sbjct 1 MAATKFTAIYINNEGKIIKREIPGMNSYKIAEKFAMMLNDPEETKLVCVIETWKMYPNNY 60
Query 61 ekktnknnLDSK 72
EK ++NL SK
Sbjct 61 EKTEKEDNLGSK 72
> Alpavirinae_Human_feces_A_047_Microviridae_AG0318_hypothetical.protein
Length=66
Score = 104 bits (260), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 48/57 (84%), Positives = 54/57 (95%), Gaps = 0/57 (0%)
Query 1 MAATKFTAIYVNNDGKLIEKEIPGMNTYKIAEKFAIMLNDTEETKVVCVIESWKLYP 57
MAATKFTAIY +N+GK+IE+EIPGMNTYKIAEKFA MLND EETK+VCV+ESWKLYP
Sbjct 1 MAATKFTAIYADNNGKIIEREIPGMNTYKIAEKFAKMLNDPEETKLVCVVESWKLYP 57
> Alpavirinae_Human_feces_B_020_Microviridae_AG0356_hypothetical.protein
Length=55
Score = 42.7 bits (99), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 27/54 (50%), Positives = 40/54 (74%), Gaps = 1/54 (2%)
Query 20 KEIPGMNTYKIAEKFA-IMLNDTEETKVVCVIESWKLYPnenekktnknnLDSK 72
KE+PG +TYK+A A ++ + E ++V V+ESWKLYPNE ++KTN +NLD +
Sbjct 2 KELPGKHTYKVAVTVAKALMQEVEGEEIVAVVESWKLYPNETKEKTNNDNLDGE 55
> Gokush_Human_feces_C_014_Microviridae_AG0150_putative.VP2
Length=286
Score = 19.6 bits (39), Expect = 4.6, Method: Composition-based stats.
Identities = 8/31 (26%), Positives = 16/31 (52%), Gaps = 0/31 (0%)
Query 24 GMNTYKIAEKFAIMLNDTEETKVVCVIESWK 54
G NT ++A+K+ + K + +SW+
Sbjct 180 GNNTAELADKYLQLAKQATSMKQIKAAKSWE 210
> Gokush_Human_feces_B_068_Microviridae_AG0329_putative.AF306496_8.Vp5
Length=97
Score = 18.9 bits (37), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 6/16 (38%), Positives = 11/16 (69%), Gaps = 0/16 (0%)
Query 8 AIYVNNDGKLIEKEIP 23
+Y ++ G+LI E+P
Sbjct 55 GLYDSDTGRLIVDEVP 70
> Gokush_Human_feces_E_009_Microviridae_AG0283_putative.VP2
Length=294
Score = 18.9 bits (37), Expect = 8.7, Method: Composition-based stats.
Identities = 8/26 (31%), Positives = 13/26 (50%), Gaps = 0/26 (0%)
Query 12 NNDGKLIEKEIPGMNTYKIAEKFAIM 37
NN+ L ++ N +AEK+ M
Sbjct 152 NNENALKIADVNAKNNLAVAEKYTAM 177
Lambda K H a alpha
0.316 0.132 0.377 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3698730