bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-35_CDS_annotation_glimmer3.pl_2_4
Length=360
Score E
Sequences producing significant alignments: (Bits) Value
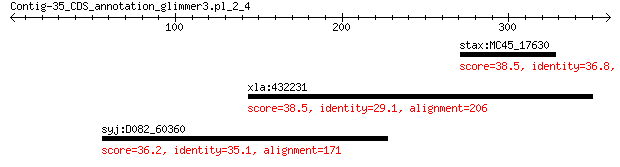
stax:MC45_17630 exopolysaccharide biosynthesis protein 38.5 2.6
xla:432231 myh15, myh7, vMHC; myosin, heavy chain 15 38.5
syj:D082_60360 hypothetical protein 36.2 8.6
> stax:MC45_17630 exopolysaccharide biosynthesis protein
Length=738
Score = 38.5 bits (88), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 21/57 (37%), Positives = 38/57 (67%), Gaps = 3/57 (5%)
Query 271 AKVETDYKRMLTKVEDSKLEPTLKEIQARIDNLDKSSLSLEQRRILDSYETEIRAAL 327
A++ +Y+R++T+ E S P + +Q++ID LD+ S+S E+RR S E + R+A+
Sbjct 318 AELRAEYQRLMTQFEPSY--PAAQAVQSQIDQLDR-SISTEERRATGSLEADYRSAV 371
> xla:432231 myh15, myh7, vMHC; myosin, heavy chain 15
Length=1937
Score = 38.5 bits (88), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 60/214 (28%), Positives = 99/214 (46%), Gaps = 15/214 (7%)
Query 144 ADADIALKDAQRIKTLAEAQGQTNENSLFEFARSAAESDALSKRFRLELDKVNAEFAQAF 203
++ + +L + +++K E + + +L E + ++ + RF+LEL ++ AEF +
Sbjct 1519 SEGNKSLHEIEKVKKQVEQEKSEVQVALEEAEGALEHEESKTLRFQLELAQIKAEFDRKL 1578
Query 204 AEADLAQRKAL--LQEIWSRVKSNLAQAA--RTDADRLTIDTLRDLQAKSLGAGISLTQT 259
AE D LQ I ++SNL A R +A RL DL I L+ +
Sbjct 1579 AEKDEETENVRRNLQRIIESLQSNLESEAKSRNEAVRLKKKMEGDLNE----MEIQLSHS 1634
Query 260 QTSATQAQTDLAKVETDYKRMLTKVEDS-KLEPTLKE---IQARIDNLDKSSLSLEQRRI 315
AT+A L ++ YK + + +D+ L LKE + R +NL +S L E R
Sbjct 1635 TRQATEATKALRNLQAQYKDLQVQYDDTLHLSEDLKEQAAVIERRNNLLQSELD-ELRAA 1693
Query 316 LDSYETEIRAALANHEKGEMREFLRKVETTGTSL 349
LD +TE L+ E E E + + + TSL
Sbjct 1694 LD--QTERARKLSEQELLESTERVNLLHSQNTSL 1725
> syj:D082_60360 hypothetical protein
Length=264
Score = 36.2 bits (82), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 60/191 (31%), Positives = 84/191 (44%), Gaps = 38/191 (20%)
Query 56 RQKDLAQWAYTNFDSPAAQRAAYAAAG-VNPFVEGSSIQPAQVNPGSVQAAQGGDVPGSG 114
RQK+L A +NF SP Q Y+ G V+ VE P ++ G+V AA
Sbjct 78 RQKNLDSIAISNF-SPFLQVFLYSTLGPVSNGVEAPVNNPFRLILGNVAAA--------- 127
Query 115 PYQMNPMSALQSGASSII---QNAMLDRQMKNADA--DIALKDAQRIKTLAEAQGQTNEN 169
L G S+ Q A D Q + A A DI +K A+ + AE Q + E
Sbjct 128 --------FLGQGLGSLFNWSQYAGNDSQQQRAIAIGDIQIKIAELQRNRAELQQKLREQ 179
Query 170 ------SLFEFARSAAESDALSKRF--RLELDKVNAEFAQAFAEADLAQ------RKALL 215
L E AR+ A++KR RLE+ KV+ F + +E LAQ +KA
Sbjct 180 VTVEVLKLEELARAFQIEQAIAKRDKQRLEIAKVSYRFGEGDSERYLAQLSSYDRQKAAT 239
Query 216 QEIWSRVKSNL 226
WSR++S +
Sbjct 240 FREWSRLRSQV 250
Lambda K H a alpha
0.311 0.124 0.330 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 681980606862