bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-34_CDS_annotation_glimmer3.pl_2_3
Length=90
Score E
Sequences producing significant alignments: (Bits) Value
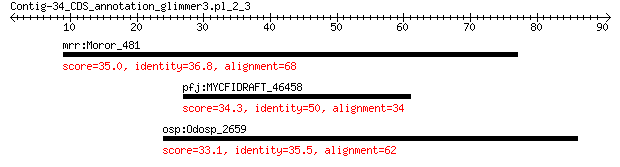
mrr:Moror_481 hypothetical protein 35.0 1.3
pfj:MYCFIDRAFT_46458 hypothetical protein 34.3 2.2
osp:Odosp_2659 hypothetical protein 33.1 5.8
> mrr:Moror_481 hypothetical protein
Length=426
Score = 35.0 bits (79), Expect = 1.3, Method: Composition-based stats.
Identities = 25/72 (35%), Positives = 31/72 (43%), Gaps = 5/72 (7%)
Query 9 WAYGQDVVKVTCIKQGEDDKFVLTVGQYSVTP----IIFDTQEQAETFLETKFKLTNFDL 64
W GQ V V +GED K ++ YS P +FD A LE F L N D
Sbjct 310 WCEGQVRVHVDWAAKGEDGKPIVQSAAYSTVPNEAFKVFDLSRPAH-ILEIYFLLRNIDQ 368
Query 65 AVIGAMCQRLNE 76
GA +R+ E
Sbjct 369 WTTGAFKERVTE 380
> pfj:MYCFIDRAFT_46458 hypothetical protein
Length=437
Score = 34.3 bits (77), Expect = 2.2, Method: Composition-based stats.
Identities = 17/34 (50%), Positives = 22/34 (65%), Gaps = 0/34 (0%)
Query 27 DKFVLTVGQYSVTPIIFDTQEQAETFLETKFKLT 60
DK VLTVG YS T + F +Q QA ++ T +LT
Sbjct 213 DKIVLTVGAYSDTLLDFKSQLQATAYVVTHVRLT 246
> osp:Odosp_2659 hypothetical protein
Length=531
Score = 33.1 bits (74), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 30/62 (48%), Gaps = 14/62 (23%)
Query 24 GEDDKFVLTVGQYSVTPIIFDTQEQAETFLETKFKLTNFDLAVIGAMCQRLNELNEQNKL 83
G D FV+ GQY+VT I +D Q AE LA+ G + R + + NK+
Sbjct 219 GNGDMFVIDPGQYNVTTIGYDPQIGAEL------------LAITGKIYPR--SMQQDNKV 264
Query 84 YS 85
YS
Sbjct 265 YS 266
Lambda K H a alpha
0.315 0.131 0.371 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 128026330890