bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-29_CDS_annotation_glimmer3.pl_2_1
Length=295
Score E
Sequences producing significant alignments: (Bits) Value
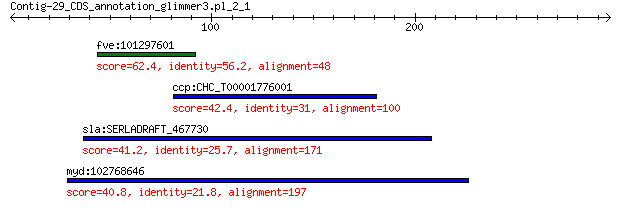
fve:101297601 minor spike protein H-like 62.4 3e-09
ccp:CHC_T00001776001 hypothetical protein 42.4 0.086
sla:SERLADRAFT_467730 glycoside hydrolase family 17 protein 41.2 0.19
myd:102768646 inactive dipeptidyl peptidase 10-like 40.8 0.24
> fve:101297601 minor spike protein H-like
Length=139
Score = 62.4 bits (150), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 27/48 (56%), Positives = 40/48 (83%), Gaps = 0/48 (0%)
Query 44 EAQKNRDWQERMSSTAHQREVRDLIAAGLNPVLSVTGGSGAAVTSGAT 91
EAQ+NR++QER+S++A+QR+V DL +AGLNP+L+ G GA+ SG+T
Sbjct 25 EAQRNREFQERLSNSAYQRQVADLSSAGLNPMLAYIKGGGASTPSGST 72
> ccp:CHC_T00001776001 hypothetical protein
Length=595
Score = 42.4 bits (98), Expect = 0.086, Method: Compositional matrix adjust.
Identities = 31/102 (30%), Positives = 54/102 (53%), Gaps = 4/102 (4%)
Query 81 GSGAAVTSGATASSSAPSGAMGSVDNSATGAVAGLFGSLLSSFLSLEGTRVSAQSNQAIA 140
G+G + S +T+++ AP+G VD A + +F SLL SLEG + + ++ I
Sbjct 342 GAGGTLVSTSTSAAVAPTGR--GVDGDAGFDLTTMFHSLLDLERSLEGKYFAPRMSRDIM 399
Query 141 DKYTA--MSKYTSELQAQTQLTSTNIQAMAQKYTADAHLAGT 180
+ +T +++ T E++A QL T M ++ TA + A T
Sbjct 400 NPHTGEIVARTTGEMRASLQLAKTKDARMLRELTAQTYYAIT 441
> sla:SERLADRAFT_467730 glycoside hydrolase family 17 protein
Length=480
Score = 41.2 bits (95), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 44/188 (23%), Positives = 78/188 (41%), Gaps = 26/188 (14%)
Query 37 ARKYNSQEAQKNRDWQERMSSTAHQREVRDLIAA---GLNPVLSVTGGSGAAVTSGATAS 93
AR Y + +A N W E+ + R + ++A GL ++++ G ++ +++
Sbjct 91 ARSYENLDAPPNNQWLEKQPQSQGSRRAKWIVAGSLIGLAALIAIGVALGVTLSKKHSSN 150
Query 94 SSAPSGAMGSVDNSATGAVAGLFGSLLSSFLSLEGTRVSAQSNQAIADKYTAMSKYTSEL 153
SSAPSG + N+ A L+ SF L T + +Q + D ++ + ++
Sbjct 151 SSAPSGTNPNTPNNFPKNNA-----LIQSFYGLAYTPLGSQ----LPDCGNSIDEVIEDI 201
Query 154 QAQTQLT-------------STNIQAMAQ-KYTADAHLAGTKYAADQSAAAQKVSASIHA 199
Q +QLT S + A+ Q + +L A D A ++ I
Sbjct 202 QLMSQLTTRIRLYGADCNQSSLVLSAIQQTQVNMSVYLGNYPSATDNGTAYERQRGEIQV 261
Query 200 AAQKYGYN 207
A Q YG N
Sbjct 262 ALQNYGAN 269
> myd:102768646 inactive dipeptidyl peptidase 10-like
Length=448
Score = 40.8 bits (94), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 43/197 (22%), Positives = 74/197 (38%), Gaps = 10/197 (5%)
Query 29 WQEQQAELARKYNSQEAQKNRDWQERMSSTAHQREVRDLIAAGLNPVLSVTGGSGAAVTS 88
WQE L+ KY+ + + + +S+TAH+ + PV S T + S
Sbjct 6 WQEATLVLSNKYSVSVSATDHKYLVHVSATAHKYSI---------PV-SATARKYSVPMS 55
Query 89 GATASSSAPSGAMGSVDNSATGAVAGLFGSLLSSFLSLEGTRVSAQSNQAIADKYTAMSK 148
S P A + + A+A + +S+ VSA +++ + K
Sbjct 56 ATDHKYSVPVSATAHKYSVSMSAIAHKYLVPVSATAHKYSVPVSATAHKYLVPVSATAHK 115
Query 149 YTSELQAQTQLTSTNIQAMAQKYTADAHLAGTKYAADQSAAAQKVSASIHAAAQKYGYNV 208
Y+ + A + A KY+ KY SA A K S + A A K+ V
Sbjct 116 YSVPVSATAHKYLIPVSATDHKYSVPVSATAHKYLVPVSATAHKYSVPMSATAHKHSVPV 175
Query 209 QSMTQRDIAAFNAQVNK 225
+ + + +A +K
Sbjct 176 SATDHKYLVPVSATAHK 192
Lambda K H a alpha
0.309 0.120 0.330 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 495615730601