bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-26_CDS_annotation_glimmer3.pl_2_7
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
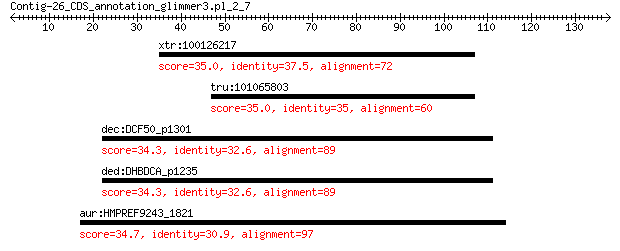
xtr:100126217 huwe1, arf-bp1, hecth9, hspc272, ib772, lasu1, m... 35.0 4.2
tru:101065803 E3 ubiquitin-protein ligase HUWE1-like 35.0 4.3
dec:DCF50_p1301 putative NADPH-dependent FMN reductase 34.3 4.5
ded:DHBDCA_p1235 putative NADPH-dependent FMN reductase 34.3 4.5
aur:HMPREF9243_1821 purA; adenylosuccinate synthase (EC:6.3.4.4) 34.7 5.0
> xtr:100126217 huwe1, arf-bp1, hecth9, hspc272, ib772, lasu1,
mule, ureb1; HECT, UBA and WWE domain containing 1, E3 ubiquitin
protein ligase
Length=4324
Score = 35.0 bits (79), Expect = 4.2, Method: Composition-based stats.
Identities = 27/72 (38%), Positives = 35/72 (49%), Gaps = 4/72 (6%)
Query 35 RYQDKFTEETYRAASGIKLALPTYYKQKLWTDQERESLRIIKEEQQVKYYNKTPIKVETL 94
R D FTE A S I++ALP +D E E+LRI K V+ TPIK TL
Sbjct 1876 RNPDIFTEV---ANSCIRIALPAPRGSGTASDDEFENLRI-KGPNAVQLVKTTPIKPSTL 1931
Query 95 EQYKEYVNAVMY 106
+ + V+Y
Sbjct 1932 PVIPDTIKEVIY 1943
> tru:101065803 E3 ubiquitin-protein ligase HUWE1-like
Length=4424
Score = 35.0 bits (79), Expect = 4.3, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 32/60 (53%), Gaps = 1/60 (2%)
Query 47 AASGIKLALPTYYKQKLWTDQERESLRIIKEEQQVKYYNKTPIKVETLEQYKEYVNAVMY 106
A+S +++ALP +D E E+LRI K V+ TP+K+ TL E + V+Y
Sbjct 1933 ASSCVRIALPAPRGAGTASDDEFENLRI-KGPNAVQLVKTTPLKLSTLPPIPETIKDVIY 1991
> dec:DCF50_p1301 putative NADPH-dependent FMN reductase
Length=166
Score = 34.3 bits (77), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 40/106 (38%), Gaps = 17/106 (16%)
Query 22 GIGYINKNSLNKHRYQDKFTEET---YRAASGIKLALPTYYKQKLWTDQERESLRIIKEE 78
G GY NK+ N+ ++D ET R A GI LA PTY+ T +
Sbjct 7 GCGYFNKSQGNQCVFKDDAVNETAARMREADGIILASPTYFGGIAGTTKAFLDRVFFSSR 66
Query 79 QQVKYYNKTPIKV----------ETLEQYKEYVNAVM----YWQSV 110
+Y T + V L+ Y E +M YW +V
Sbjct 67 AYFRYKVATAVSVVRRAGGVDVYRQLQNYLELSETIMPPSQYWMTV 112
> ded:DHBDCA_p1235 putative NADPH-dependent FMN reductase
Length=166
Score = 34.3 bits (77), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 40/106 (38%), Gaps = 17/106 (16%)
Query 22 GIGYINKNSLNKHRYQDKFTEET---YRAASGIKLALPTYYKQKLWTDQERESLRIIKEE 78
G GY NK+ N+ ++D ET R A GI LA PTY+ T +
Sbjct 7 GCGYFNKSQGNQCVFKDDAVNETAARMREADGIILASPTYFGGIAGTTKAFLDRVFFSSR 66
Query 79 QQVKYYNKTPIKV----------ETLEQYKEYVNAVM----YWQSV 110
+Y T + V L+ Y E +M YW +V
Sbjct 67 AYFRYKVATAVSVVRRAGGVDVYRQLQNYLELSETIMPPSQYWMTV 112
> aur:HMPREF9243_1821 purA; adenylosuccinate synthase (EC:6.3.4.4)
Length=428
Score = 34.7 bits (78), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 30/98 (31%), Positives = 45/98 (46%), Gaps = 27/98 (28%)
Query 17 TSKGIGIGYINKNSLNKHRYQDKFTEETYRAASGIKLALPTYYKQKLWTDQERESLRI-I 75
T KGIG Y++K + N R D ET+ ER S++I +
Sbjct 128 TQKGIGPAYMDKIARNGIRMADLIDPETF---------------------AERLSVQIQV 166
Query 76 KEEQQVKYYNKTPIKVETLEQYKEYVNAVMYWQSVKKY 113
K E K Y++ P+ +T+ Y+EY + Y Q +KKY
Sbjct 167 KNELLTKVYDEEPLDYDTI--YQEY---LAYGQKLKKY 199
Lambda K H a alpha
0.312 0.130 0.374 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 128430569155