bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-23_CDS_annotation_glimmer3.pl_2_6
Length=70
Score E
Sequences producing significant alignments: (Bits) Value
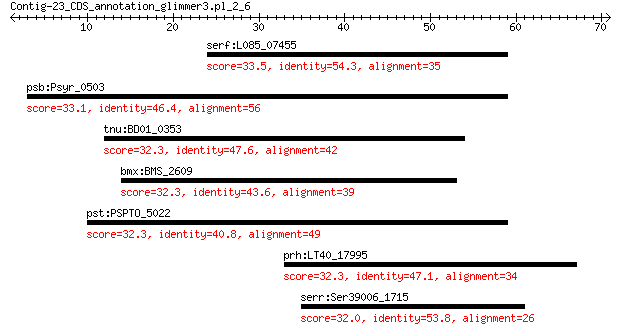
serf:L085_07455 RHS repeat-associated core domain-containing p... 33.5 3.1
psb:Psyr_0503 hypothetical protein 33.1 3.6
tnu:BD01_0353 bacitracin resistance protein BacA-like protein 32.3 5.7
bmx:BMS_2609 lipA; lipoyl synthase 32.3 6.3
pst:PSPTO_5022 hypothetical protein 32.3 7.2
prh:LT40_17995 hypothetical protein 32.3 8.1
serr:Ser39006_1715 Cysteine synthase (EC:2.5.1.47) 32.0 9.3
> serf:L085_07455 RHS repeat-associated core domain-containing
protein
Length=967
Score = 33.5 bits (75), Expect = 3.1, Method: Composition-based stats.
Identities = 19/36 (53%), Positives = 21/36 (58%), Gaps = 1/36 (3%)
Query 24 KRFTVIFRFRTYEQASDCLRF-LLENYPGIYFDIKD 58
K + R + Y QASD LR LLEN GIY DI D
Sbjct 803 KYYEETVRQKKYAQASDLLRLHLLENEGGIYKDIDD 838
> psb:Psyr_0503 hypothetical protein
Length=1044
Score = 33.1 bits (74), Expect = 3.6, Method: Composition-based stats.
Identities = 26/69 (38%), Positives = 32/69 (46%), Gaps = 14/69 (20%)
Query 3 FDYFHR---------FELAYSPFFVRKRVGKRFTVIFRF---RTYEQASDCLRF-LLENY 49
FDYF E A+ P F+ G+ F FR R Y ASD LR+ L++ Y
Sbjct 811 FDYFFEQPQMHISRLKEEAFYPDFLSGENGEAFNY-FRHSDNRNYAAASDILRYRLIDEY 869
Query 50 PGIYFDIKD 58
GIY D D
Sbjct 870 GGIYVDCDD 878
> tnu:BD01_0353 bacitracin resistance protein BacA-like protein
Length=257
Score = 32.3 bits (72), Expect = 5.7, Method: Composition-based stats.
Identities = 20/42 (48%), Positives = 23/42 (55%), Gaps = 5/42 (12%)
Query 12 AYSPFFVRKRVGKRFTVIFRFRTYEQASDCLRFLLENYPGIY 53
YS F V +G F V+FRFR E+ S R LLE GIY
Sbjct 35 GYSSFLVPAYLGVTFAVLFRFR--ERFS---RLLLEAMKGIY 71
> bmx:BMS_2609 lipA; lipoyl synthase
Length=323
Score = 32.3 bits (72), Expect = 6.3, Method: Composition-based stats.
Identities = 17/43 (40%), Positives = 26/43 (60%), Gaps = 4/43 (9%)
Query 14 SPFFVRKRVG--KRFTVIFRFRT--YEQASDCLRFLLENYPGI 52
+PF + + V KR T + R R YEQ+ +CL++ +NYP I
Sbjct 184 NPFVIAQNVETVKRLTHVVRDRRAGYEQSLNCLKYYKDNYPHI 226
> pst:PSPTO_5022 hypothetical protein
Length=1042
Score = 32.3 bits (72), Expect = 7.2, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 25/52 (48%), Gaps = 3/52 (6%)
Query 10 ELAYSPFFVRKRVGKRFTVIFRF--RTYEQASDCLRF-LLENYPGIYFDIKD 58
E A+ P F+ G+ F R Y ASD LR+ L+ Y GIY D D
Sbjct 825 EEAFFPKFLSGENGEAFNYFMHSDNRNYAAASDILRYRLINEYGGIYLDCDD 876
> prh:LT40_17995 hypothetical protein
Length=1027
Score = 32.3 bits (72), Expect = 8.1, Method: Composition-based stats.
Identities = 16/35 (46%), Positives = 22/35 (63%), Gaps = 1/35 (3%)
Query 33 RTYEQASDCLRF-LLENYPGIYFDIKDVSFSHLDK 66
+ Y ASD LR+ LL+ Y GIY D+ + + LDK
Sbjct 828 KNYAAASDLLRYPLLDTYGGIYTDVDNTFVAVLDK 862
> serr:Ser39006_1715 Cysteine synthase (EC:2.5.1.47)
Length=354
Score = 32.0 bits (71), Expect = 9.3, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 20/28 (71%), Gaps = 2/28 (7%)
Query 35 YEQASDC--LRFLLENYPGIYFDIKDVS 60
Y++++D +R L NYPGIYF +KD S
Sbjct 17 YQRSADTHLIRVTLSNYPGIYFYLKDES 44
Lambda K H a alpha
0.334 0.147 0.453 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 126668436524