bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-23_CDS_annotation_glimmer3.pl_2_1
Length=362
Score E
Sequences producing significant alignments: (Bits) Value
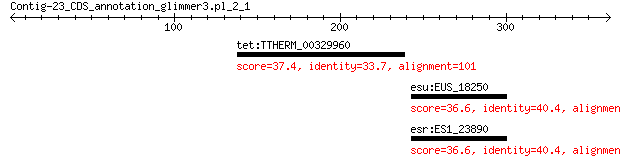
tet:TTHERM_00329960 TPR Domain containing protein 37.4 6.5
esu:EUS_18250 carbohydrate ABC transporter substrate-binding p... 36.6 8.0
esr:ES1_23890 carbohydrate ABC transporter substrate-binding p... 36.6 8.6
> tet:TTHERM_00329960 TPR Domain containing protein
Length=1280
Score = 37.4 bits (85), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 34/111 (31%), Positives = 58/111 (52%), Gaps = 17/111 (15%)
Query 138 TKI--SLNHLKIITESSIFQLIINRIG---QSYGSIQTCTRSRFSDSCFSKS-CLSLHKT 191
TKI +NH +II+ L+ N +G +SY IQ ++R +S ++ C + H
Sbjct 239 TKILGKINHPQIISSQENIALVQNELGKFQESYSLIQEVVKNR--ESLYTSDICPNSHPQ 296
Query 192 FTQIFIQSLGPIPISLSIIKRS----RRTLTLLLSIFQLNIRIRRRIPSQS 238
+ Q+LG I L ++K+S ++ L+L SI+Q + R+I SQ+
Sbjct 297 IADAY-QNLGTIQQDLGMLKKSLNNQKKALSLRESIYQYD----RQIQSQN 342
> esu:EUS_18250 carbohydrate ABC transporter substrate-binding
protein, CUT1 family (TC 3.A.1.1.-)
Length=489
Score = 36.6 bits (83), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query 243 TGTRSNRTMSRGSTTTGS---RNSSRNTNSRLAKNSSRIQTSRKITLKNSSRIIIRILII 299
G SN T S GS+TT S NSS+ +S +SS TS K TLK + I +
Sbjct 20 AGCSSNATSSAGSSTTDSSKTENSSKTEDSSKTDDSSNAGTSDKFTLKEGEGKTLNIAVW 79
> esr:ES1_23890 carbohydrate ABC transporter substrate-binding
protein, CUT1 family (TC 3.A.1.1.-)
Length=489
Score = 36.6 bits (83), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query 243 TGTRSNRTMSRGSTTTGS---RNSSRNTNSRLAKNSSRIQTSRKITLKNSSRIIIRILII 299
G SN T S GS+TT S NSS+ +S +SS TS K TLK + I +
Sbjct 20 AGCSSNATSSAGSSTTDSSKTENSSKTEDSSKTDDSSNAGTSDKFTLKEGEGKTLNIAVW 79
Lambda K H a alpha
0.326 0.136 0.376 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 688208740258