bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-22_CDS_annotation_glimmer3.pl_2_5
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
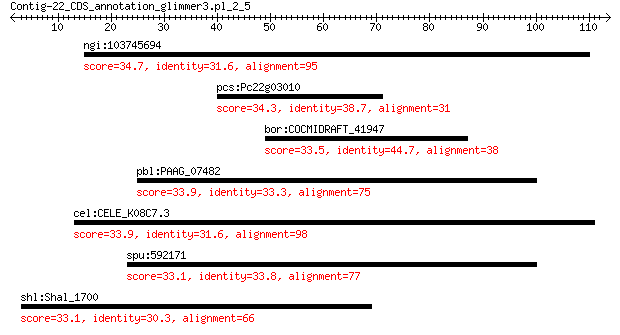
ngi:103745694 Lama3; laminin, alpha 3 34.7
pcs:Pc22g03010 hypothetical protein 34.3 2.8
bor:COCMIDRAFT_41947 hypothetical protein 33.5 4.9
pbl:PAAG_07482 ankyrin repeat protein nuc-2 33.9 6.0
cel:CELE_K08C7.3 epi-1; Protein EPI-1, isoform C 33.9 6.2
spu:592171 heat shock 70 kDa protein IV-like 33.1 7.9
shl:Shal_1700 FAD dependent oxidoreductase 33.1 7.9
> ngi:103745694 Lama3; laminin, alpha 3
Length=3237
Score = 34.7 bits (78), Expect = 2.8, Method: Composition-based stats.
Identities = 30/107 (28%), Positives = 50/107 (47%), Gaps = 21/107 (20%)
Query 15 EVLQETAPID----------IQQEIESYADECD--IKSIIRKASFDPQFLKSLSEGAMTG 62
E LQE A I+ + + I+S A E D IK++IR LK +S G
Sbjct 1904 ETLQEKAQINSRKAQTLYNNVDRTIQS-AKELDTKIKTVIRNVHI---LLKQISRPDGEG 1959
Query 63 AEVDITEWPQNIHEYHRMIATAQANAMKLEELQKTATEEPKAETKTK 109
++ + +W + + E HRMI ++ + +K TE A+T+ +
Sbjct 1960 NDLPLGDWSKELAEAHRMI-----RVLRDRDFRKQLTEAEAAKTEAQ 2001
> pcs:Pc22g03010 hypothetical protein
Length=168
Score = 34.3 bits (77), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 12/31 (39%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 40 SIIRKASFDPQFLKSLSEGAMTGAEVDITEW 70
S+ R FDPQ++ ++ G++TGA V W
Sbjct 126 SLGRNPGFDPQYIPVMTVGSLTGARVSSESW 156
> bor:COCMIDRAFT_41947 hypothetical protein
Length=206
Score = 33.5 bits (75), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 17/39 (44%), Positives = 25/39 (64%), Gaps = 1/39 (3%)
Query 49 PQFLKSLSEGAMTGAEVDITEWPQN-IHEYHRMIATAQA 86
P+FL LS+G +G EV++T+ N I YH + A A+A
Sbjct 65 PEFLTKLSKGQPSGIEVEVTDLETNSISTYHAIKAAARA 103
> pbl:PAAG_07482 ankyrin repeat protein nuc-2
Length=1036
Score = 33.9 bits (76), Expect = 6.0, Method: Composition-based stats.
Identities = 25/82 (30%), Positives = 45/82 (55%), Gaps = 10/82 (12%)
Query 25 IQQEIESYADECDIKSIIRKASFDPQFLKSLSEGAMTGAEVDITEWPQ--NIH-----EY 77
+QQ +E +E + I++K F+ + L+ LS+ A T + +D+ W + NI
Sbjct 162 LQQFVE--INETAVSKILKKPCFNREVLRDLSDRATT-SRLDLEAWAEGENIQFDASRLA 218
Query 78 HRMIATAQANAMKLEELQKTAT 99
R++ATA+ + L+ LQ T+T
Sbjct 219 DRVVATAEEDDSDLQILQTTST 240
> cel:CELE_K08C7.3 epi-1; Protein EPI-1, isoform C
Length=3683
Score = 33.9 bits (76), Expect = 6.2, Method: Composition-based stats.
Identities = 31/101 (31%), Positives = 42/101 (42%), Gaps = 13/101 (13%)
Query 13 GQEVLQETA--PIDIQQEIESYAD-ECDIKSIIRKASFDPQFLKSLSEGAMTGAEVDITE 69
G E LQE +Q + S A+ I S + + DP+ LK E MT +
Sbjct 2247 GTEFLQEVMKRAQRARQSVRSLAEIALAIGSSSKAVNVDPRLLKEAEETLMTLEAASADQ 2306
Query 70 WPQNIHEYHRMIATAQANAMKLEELQKTATEEPKAETKTKE 110
+P+ AQ KLEE+QK EE + K KE
Sbjct 2307 YPEK----------AQTVPGKLEEIQKKIQEETEKLDKQKE 2337
> spu:592171 heat shock 70 kDa protein IV-like
Length=154
Score = 33.1 bits (74), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 26/77 (34%), Positives = 40/77 (52%), Gaps = 8/77 (10%)
Query 23 IDIQQEIESYADECDIKSIIRKASFDPQFLKSLSEGAMTGAEVDITEWPQNIHEYHRMIA 82
I ++ ++ESYA ++KS I AS + + S E +T A D+ W N +A
Sbjct 51 IAVRNQLESYA--FNVKSAINDASVESKLSSSDKE-VVTKAVDDVITWMDN-----NSLA 102
Query 83 TAQANAMKLEELQKTAT 99
+ + KLEELQKT +
Sbjct 103 NKEEFSFKLEELQKTCS 119
> shl:Shal_1700 FAD dependent oxidoreductase
Length=429
Score = 33.1 bits (74), Expect = 7.9, Method: Composition-based stats.
Identities = 20/70 (29%), Positives = 33/70 (47%), Gaps = 12/70 (17%)
Query 3 KQYVWTKDEKGQEVLQETAPIDIQQEIESYADECDIK----SIIRKASFDPQFLKSLSEG 58
+QYVW +G E+++E + Y +CD+K KAS + K+ SEG
Sbjct 97 EQYVWNMRWRGHEIIKE--------RVNKYNIDCDLKFGHIHTAYKASHLKELEKTYSEG 148
Query 59 AMTGAEVDIT 68
G E +++
Sbjct 149 CKRGMEAELS 158
Lambda K H a alpha
0.307 0.123 0.338 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 126991827136