bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-21_CDS_annotation_glimmer3.pl_2_6
Length=238
Score E
Sequences producing significant alignments: (Bits) Value
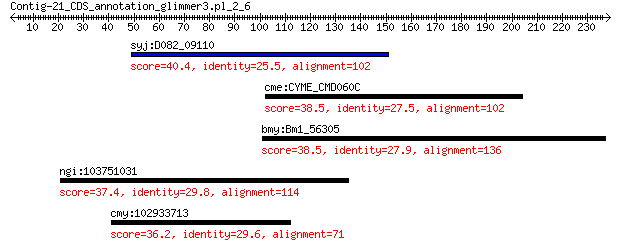
syj:D082_09110 Transcriptional regulator PchR 40.4 0.19
cme:CYME_CMD060C 20S core proteasome subunit beta 7 38.5
bmy:Bm1_56305 leucyl aminopeptidase 38.5 1.0
ngi:103751031 Dchs2; dachsous cadherin-related 2 37.4
cmy:102933713 FURIN; furin (paired basic amino acid cleaving e... 36.2 6.8
> syj:D082_09110 Transcriptional regulator PchR
Length=329
Score = 40.4 bits (93), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 26/111 (23%), Positives = 49/111 (44%), Gaps = 14/111 (13%)
Query 49 KRWWKKYKEPLKHW-----LITELGHDNTKRIHLHGIIWTELTEEQFEKEWGYGWIFFGY 103
+R+ + ++P+ ++ L T LGH +T+R L IW +L +++F + W
Sbjct 21 ERFEGQARQPVDNYETNCRLPTYLGHGHTRRFELSSGIWLDLIDKEFTQPWA-----LKM 75
Query 104 EVNERTINY--IIKYITKRDEANPEFNGKI--FTSKGIGKEYIGENSLRRH 150
+E + + ++ DE P K+ F+ GI Y+ RH
Sbjct 76 PAHEHLVQFTILLSGAVDYDETYPTLGAKMGYFSGSGISPGYVARYGRLRH 126
> cme:CYME_CMD060C 20S core proteasome subunit beta 7
Length=222
Score = 38.5 bits (88), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 28/104 (27%), Positives = 47/104 (45%), Gaps = 9/104 (9%)
Query 102 GYEVNERTI-NYIIKYI-TKRDEANPEFNGKIFTSKGIGKEYIGENSLRRHRYQDRFTEE 159
GYE++ R + +Y+ + + +R +P +N KG ++G L Y F E
Sbjct 87 GYELSPRALHSYLARVLYGRRSRMDPLYNSLAVAGKG---RFLGCIDL----YGTAFESE 139
Query 160 TYRTNSGIKIALPTYYKQKLWTVQEREALRIIKEEKQVKYYNKT 203
T G +ALP KQ + E +R+++E +V YY
Sbjct 140 AVATGIGALLALPLIRKQWHAALAMDEGVRLLEECLRVLYYRDC 183
> bmy:Bm1_56305 leucyl aminopeptidase
Length=406
Score = 38.5 bits (88), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 38/159 (24%), Positives = 62/159 (39%), Gaps = 28/159 (18%)
Query 101 FGYEVNERTINYIIKYITKRDEANPEFNGKIFTSKGIGKEYIGENSLRR----------- 149
FG + E +I+YITK D A + G G +G +SL
Sbjct 28 FGNDTTE-----LIRYITKGDGAGLAYQWLSTLVDGFGHRMVGSDSLEEAIDFLAKSLEE 82
Query 150 HRYQDRFTEET------YRTNSGIKIALPTYYKQKLWTVQEREALRI------IKEEKQV 197
+ D TEE R + ++I P + + + + E I I++
Sbjct 83 DDFDDVHTEEVPNLPNWVREDDNVEIIEPRHQRLNVLALGGCEPANITGEVVVIRDLDDS 142
Query 198 KYYNKTPIRVETIEQYREYVNAVKYWQSIKKYDGKRKKG 236
K+ N + V T +Q++ Y VKY QS+K ++ G
Sbjct 143 KFINVSGKIVVTAQQFKGYPQTVKYRQSVKLFESLGAIG 181
> ngi:103751031 Dchs2; dachsous cadherin-related 2
Length=2706
Score = 37.4 bits (85), Expect = 2.8, Method: Composition-based stats.
Identities = 34/115 (30%), Positives = 52/115 (45%), Gaps = 20/115 (17%)
Query 21 FSEESLKKLEYDEKEPNKAPQKAISLFRKRWWKKYKEPLKHWLITELGHDNTKRIHLHGI 80
F ++++ L E N P K +SL K PL+ H N K LH
Sbjct 753 FMSQNIRHLVIPE---NTKPAKIMSLM------KPPSPLQQ-------HHNGK---LHFS 793
Query 81 IWTELTEEQFEKEWGYGWIFFGYEVN-ERTINYIIKYITKRDEANPEFNGKIFTS 134
I+ E ++ FE + G +F E++ E T +Y+I+ I+K +P N IF S
Sbjct 794 IFAEDKDDHFEIDSSTGDLFLSEELDYEITSHYLIRVISKDHSHDPPLNSTIFLS 848
> cmy:102933713 FURIN; furin (paired basic amino acid cleaving
enzyme)
Length=787
Score = 36.2 bits (82), Expect = 6.8, Method: Composition-based stats.
Identities = 21/71 (30%), Positives = 36/71 (51%), Gaps = 11/71 (15%)
Query 41 QKAISLFRKRWWKKYKEPLKHWLITELGHDNTKRIHLHGIIWTELTEEQFEKEWGYGWIF 100
++++S R R +EP HWL ++ TKR +TE T+ +F ++W
Sbjct 75 KRSLSPHRSRHSHLAREPQLHWLEQQVAKRRTKR-----DAFTEPTDPKFPQQW------ 123
Query 101 FGYEVNERTIN 111
+ Y VN+R +N
Sbjct 124 YLYNVNQRDLN 134
Lambda K H a alpha
0.316 0.135 0.409 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 329549477264