bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-19_CDS_annotation_glimmer3.pl_2_5
Length=582
Score E
Sequences producing significant alignments: (Bits) Value
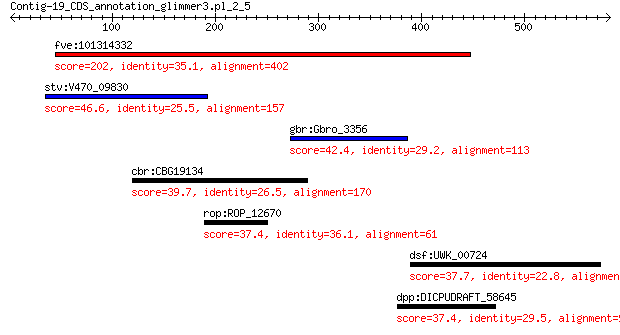
fve:101314332 capsid protein VP1-like 202 3e-55
stv:V470_09830 capsid protein 46.6 0.012
gbr:Gbro_3356 hypothetical protein 42.4 0.10
cbr:CBG19134 Hypothetical protein CBG19134 39.7 1.2
rop:ROP_12670 hypothetical protein 37.4 3.4
dsf:UWK_00724 tetratricopeptide repeat protein 37.7 7.4
dpp:DICPUDRAFT_58645 hypothetical protein 37.4 10.0
> fve:101314332 capsid protein VP1-like
Length=421
Score = 202 bits (513), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 141/407 (35%), Positives = 205/407 (50%), Gaps = 67/407 (16%)
Query 45 VLPGDTFSVDTAAIIRMTTPKYPVMDDAYIDFYYFYCPNRILWDNFKRFMGEADDAPWMP 104
VLPGDTF+V+ R+ TP +PVMD+ ++D ++F+ PNR++W+N+ +FMGE D+
Sbjct 77 VLPGDTFNVNVTMFGRLATPIFPVMDNLHLDSFFFFVPNRLVWNNWVKFMGEQDNP--AD 134
Query 105 TKTYKVPKIIIDNEEGGAVRAYPDESTILDYMGVPPKAIPVGGTGRIEINALPVRAYVKI 164
+ +Y +P+ + + GG ++ DY G+P A VG + + +ALPVRAY I
Sbjct 135 SISYSIPQQV--SPAGGYAVG-----SLQDYFGLP-TAGQVGVSNTVSHSALPVRAYNLI 186
Query 165 WNEFFRDQNVGNPAVLNTGDEDEAYRAGSGNESEVSEEKILEYAHIGGYCLPVNRFHDYF 224
+N++FRD+N+ N V++ GD G ++ S L L + HDYF
Sbjct 187 YNQWFRDENLQNSVVVDKGD---------GPDTTPSTNYTL---------LRRGKRHDYF 228
Query 225 SSCLPYPQRG-PEVTIALTGNAPL--RAYSEKDLNNRKIGTG-FFNNEYNTGIVNHTNIS 280
+S LP+PQ+G V++ L +AP+ S D+ G N Y+TG
Sbjct 229 TSALPWPQKGGTAVSLPLGTSAPIAFSGASGSDVGVISTTQGNLIKNMYSTG-------- 280
Query 281 FTKEGTKFSVNKNNNGNTAPLVNGQYIQTMSQDDANFFDAWLGTDLSNIEAATINQLRQA 340
GT + +A + G Y DLS AATINQLRQ+
Sbjct 281 ---SGTSLKIG------SATVATGLY-----------------ADLSAATAATINQLRQS 314
Query 341 FAVQHYYEALARGGSRYREQVRALFGVSISDKTVQIPEYLGGGRYHVNMNQI-VQTSGQE 399
F +Q E ARGG+RY E +R+ FGV+ D +Q PEYLGGG +N+ I
Sbjct 315 FQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPINIAPIAQTGGTGA 374
Query 400 SNYGTPIGETGAMSVTPINESSFTKSFEEHGFVIGVMCVRHDHSYQQ 446
TP G A F++SF EHG VIG++ VR D +YQQ
Sbjct 375 QGTTTPQGNLAAFGTYMAKGHGFSQSFVEHGHVIGLVSVRADLTYQQ 421
> stv:V470_09830 capsid protein
Length=427
Score = 46.6 bits (109), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 40/166 (24%), Positives = 71/166 (43%), Gaps = 37/166 (22%)
Query 35 GKLIPFYVDEVLPGDTFSVDTAAIIRMTTPKYPVMDDAYIDFYYFYCPNRILW-DNFKRF 93
G+LI V+ GD+F +D +R++ + + D+ +D + FY P+R ++ + + +F
Sbjct 26 GRLITISTTPVIAGDSFEMDAVGALRLSPLRRGLAIDSTVDIFTFYVPHRHVYGEQWIKF 85
Query 94 MGEADDAPWMPTKTYKVPKIIIDNEEGGAVRAYPDESTILDYMGVPPKAIPVGGTGRIEI 153
M + +A +PT N G Y D + L GT +
Sbjct 86 MKDGVNATPLPTV----------NTTG-----YIDHAAFL-------------GTINPDT 117
Query 154 NALP---VRAYVKIWNEFFR-----DQNVGNPAVLNTGDEDEAYRA 191
N +P + Y+ I+N +F+ D+ NP LN D +R
Sbjct 118 NKIPKHLFQGYLNIYNNYFKAPWMPDRTEANPNELNQDDARYGFRC 163
> gbr:Gbro_3356 hypothetical protein
Length=194
Score = 42.4 bits (98), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 33/127 (26%), Positives = 56/127 (44%), Gaps = 18/127 (14%)
Query 273 IVNHTNISFTKEGTKFSVNKNNNGNTAPLVNGQ---YIQTMSQDDANFFDAWLGTD---- 325
+V H I+ T + T F+ + + TAP G + +++ A DAW D
Sbjct 41 VVGHI-IAVTDKFTSFARAETDTPRTAPAPRGDPAPLRRALAESVAASADAWPSCDGSRR 99
Query 326 -------LSNIEAATINQLRQAFAVQHYYEALARGGSRYREQVRALFGVSISDKTVQIPE 378
S ++AA+IN + A+ H ++ A G YR RA+ + + + PE
Sbjct 100 CRLPFGTFSAVQAASINMMD---ALVHAWDIAAANGIDYRMPSRAIGPAIVIARRLVTPE 156
Query 379 YLGGGRY 385
+ GG+Y
Sbjct 157 AVAGGQY 163
> cbr:CBG19134 Hypothetical protein CBG19134
Length=257
Score = 39.7 bits (91), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 45/190 (24%), Positives = 78/190 (41%), Gaps = 24/190 (13%)
Query 119 EGGAVRAYPDESTILDYMGVP-PKAIPVGGTGRIEINALP--------VRAYVKIWNE-- 167
E V + EST+ + + K P+ ++ A P VRA +K+ E
Sbjct 72 EENCVIVHASESTLDVRLELKMKKKWPLCKVVKVNSQAAPDENNQIEVVRAKLKLTFEHQ 131
Query 168 -FFRDQNVGNPAVLNTGDEDEAYRAGSGNESEVSEEKILEYAHIGGYC--LPVNRFHDYF 224
RD+N + L + Y + VS +++ +++ + PV +
Sbjct 132 NLRRDRNSVDLDTLMRVFLNSRYIEALYLDEFVSVDELFQFSEQKDFAKLFPVCQL---- 187
Query 225 SSCLPYPQRGPEVT------IALTGNAPLRAYSEKDLNNRKIGTGFFNNEYNTGIVNHTN 278
+ +P P+ + T + ++PL S + +N KI F+N EY T H +
Sbjct 188 TMVIPKPKNKSDETKFWNFILYFVVDSPLVLVSSRVVNQDKIDLVFYNPEYKTCNFKHFS 247
Query 279 ISFTKEGTKF 288
I F+KEG KF
Sbjct 248 IPFSKEGVKF 257
> rop:ROP_12670 hypothetical protein
Length=169
Score = 37.4 bits (85), Expect = 3.4, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 30/61 (49%), Gaps = 3/61 (5%)
Query 189 YRAGSGNESEVSEEKILEYAHIGGYCLPVNRFHDYFSSCLPYPQRGPEVTIALTGNAPLR 248
+ AG +E S+ + +A I CL + R+ D S PYP R +T A TG PL
Sbjct 3 FDAGLRRFNEASQSEATTWARI---CLDIPRWADELVSARPYPDRSSLLTAARTGAGPLS 59
Query 249 A 249
A
Sbjct 60 A 60
> dsf:UWK_00724 tetratricopeptide repeat protein
Length=419
Score = 37.7 bits (86), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 42/202 (21%), Positives = 81/202 (40%), Gaps = 28/202 (14%)
Query 389 MNQIVQTSGQESNYGTPIGETGAMSVTPINESSFTKSFEEHGFVIGVM-----CVRHDHS 443
+NQ+ T+G+ GT + G + I S ++S ++ + + +R
Sbjct 67 LNQVSLTAGELQKQGTRLAREGKLEEARI---SLSRSLKKDPTSLVTLNNLGLVMRKLER 123
Query 444 YQQGLERFWSRSDRLDYYFPQFANLG----EQPVK-------KKEIMLTGKSTDEETFGY 492
+ + L+ + S + Y + NLG +Q V KK +L + D G
Sbjct 124 FDEALQAYISALESDKNYALTYKNLGILLEKQGVHDQAALAYKKYCVLAPNAADAHNVGA 183
Query 493 QEAWADYRM--KPNRVSGKMRSNAEGTLDFWHYADNYATVPTLSQEWMKEGKNEIARTLI 550
+ W ++ P + + + ++ E L +D Y L EW G N A +++
Sbjct 184 RADWLQSQINTSPTQSTTQQKAEEEQEL-----SDQYMAEKAL--EWHDIGANLFAESVV 236
Query 551 VENEPQFFGAIRVMNKTTRCMP 572
++ + F AI + K T +P
Sbjct 237 EKDRDKLFSAIDYLEKATVGLP 258
> dpp:DICPUDRAFT_58645 hypothetical protein
Length=388
Score = 37.4 bits (85), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 44/100 (44%), Gaps = 9/100 (9%)
Query 376 IPEYLGGGR-----YHVNMNQIVQTSGQESNYGTPIGETGAMSVTPINESSFTKSFEEHG 430
I +LG G+ Y +N N + + ++ YG IG AM V +S+ K E
Sbjct 26 ITGFLGSGKTTFLNYILNENHGKKIAVIQNEYGQSIGIETAMIV----DSNGEKVQEWLE 81
Query 431 FVIGVMCVRHDHSYQQGLERFWSRSDRLDYYFPQFANLGE 470
F G +C + Q +E RSD+ DY + +G+
Sbjct 82 FPNGCICCTVKDDFLQSIEDLLKRSDKFDYILIESTGMGD 121
Lambda K H a alpha
0.317 0.135 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1335954814206