bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-18_CDS_annotation_glimmer3.pl_2_4
Length=93
Score E
Sequences producing significant alignments: (Bits) Value
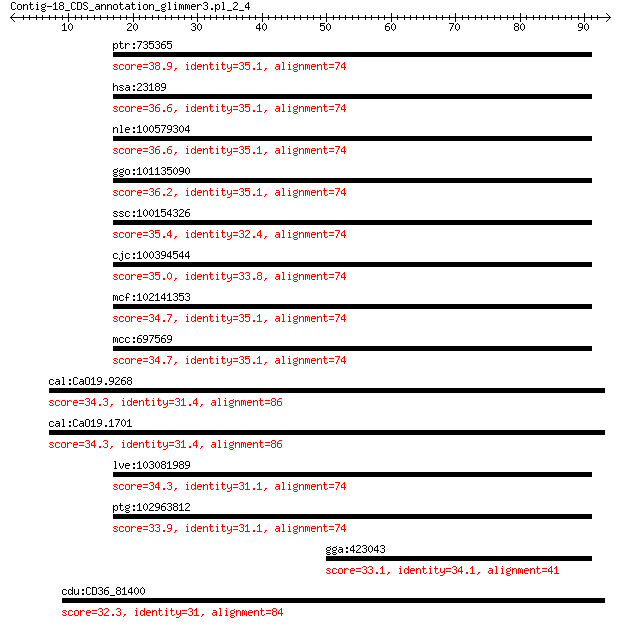
ptr:735365 KANK1, ANKRD15; KN motif and ankyrin repeat domains 1 38.9
hsa:23189 KANK1, ANKRD15, CPSQ2, KANK; KN motif and ankyrin re... 36.6 0.41
nle:100579304 KANK1; KN motif and ankyrin repeat domains 1 36.6
ggo:101135090 KANK1; KN motif and ankyrin repeat domain-contai... 36.2 0.55
ssc:100154326 KANK1; KN motif and ankyrin repeat domains 1 35.4
cjc:100394544 KANK1; KN motif and ankyrin repeat domains 1 35.0
mcf:102141353 KANK1; KN motif and ankyrin repeat domains 1 34.7
mcc:697569 KANK1; KN motif and ankyrin repeat domains 1 34.7
cal:CaO19.9268 RKI1; similar to S. cerevisiae RKI1 (YOR095C) p... 34.3 2.1
cal:CaO19.1701 RKI1; similar to S. cerevisiae RKI1 (YOR095C) p... 34.3 2.1
lve:103081989 KANK1; KN motif and ankyrin repeat domains 1 34.3
ptg:102963812 KANK1; KN motif and ankyrin repeat domains 1 33.9
gga:423043 SBF2; SET binding factor 2 33.1
cdu:CD36_81400 phosphoriboisomerase, putative (EC:5.3.1.6) 32.3 9.7
> ptr:735365 KANK1, ANKRD15; KN motif and ankyrin repeat domains
1
Length=474
Score = 38.9 bits (89), Expect = 0.069, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 40/74 (54%), Gaps = 3/74 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEADEYEKKLKRS 76
T+Q E+E + GK +++ + FPT+E +N D + IAA +YAC + K + +
Sbjct 44 TSQSEQEVGTLEGKPISSLDAFPTQEGTLSPVNLTD-DQIAAGLYACTNNESTLKSIMK- 101
Query 77 AKKVTKKIINDAKK 90
KK K N AKK
Sbjct 102 -KKDGNKDSNGAKK 114
> hsa:23189 KANK1, ANKRD15, CPSQ2, KANK; KN motif and ankyrin
repeat domains 1
Length=1352
Score = 36.6 bits (83), Expect = 0.41, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 39/74 (53%), Gaps = 3/74 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEADEYEKKLKRS 76
T+Q E+E GK +++ + FPT+E +N D + IAA +YAC + K + +
Sbjct 922 TSQPEQEVGTSEGKPISSLDAFPTQEGTLSPVNLTD-DQIAAGLYACTNNESTLKSIMK- 979
Query 77 AKKVTKKIINDAKK 90
KK K N AKK
Sbjct 980 -KKDGNKDSNGAKK 992
> nle:100579304 KANK1; KN motif and ankyrin repeat domains 1
Length=1352
Score = 36.6 bits (83), Expect = 0.49, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 39/74 (53%), Gaps = 3/74 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEADEYEKKLKRS 76
T+Q E+E GK +++ + FPT+E +N D + IAA +YAC + K + +
Sbjct 922 TSQPEQEVGTSEGKPVSSLDAFPTQEGTLSPVNLTD-DQIAAGLYACTNNESTLKSIMK- 979
Query 77 AKKVTKKIINDAKK 90
KK K N AKK
Sbjct 980 -KKDGNKDSNGAKK 992
> ggo:101135090 KANK1; KN motif and ankyrin repeat domain-containing
protein 1 isoform 1
Length=1352
Score = 36.2 bits (82), Expect = 0.55, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 39/74 (53%), Gaps = 3/74 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEADEYEKKLKRS 76
T+Q E+E GK +++ + FPT+E +N D + IAA +YAC + K + +
Sbjct 922 TSQPEQEVGTSEGKPISSLDAFPTQEGTLSPLNLTD-DQIAAGLYACTNNESTLKSIMK- 979
Query 77 AKKVTKKIINDAKK 90
KK K N AKK
Sbjct 980 -KKDGNKDSNGAKK 992
> ssc:100154326 KANK1; KN motif and ankyrin repeat domains 1
Length=539
Score = 35.4 bits (80), Expect = 0.95, Method: Composition-based stats.
Identities = 24/74 (32%), Positives = 36/74 (49%), Gaps = 3/74 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEADEYEKKLKRS 76
T+Q E+E GK ++ + FP +E +N D + IAA +Y C + K + +
Sbjct 138 TSQHEQEVGTTEGKPISGQDTFPPQETTLSAVNLTD-DQIAAGLYVCTNNESTLKSIMK- 195
Query 77 AKKVTKKIINDAKK 90
KK K N AKK
Sbjct 196 -KKDANKDSNGAKK 208
> cjc:100394544 KANK1; KN motif and ankyrin repeat domains 1
Length=1353
Score = 35.0 bits (79), Expect = 1.3, Method: Composition-based stats.
Identities = 25/74 (34%), Positives = 38/74 (51%), Gaps = 3/74 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEADEYEKKLKRS 76
T+Q E+E GK +++ + FPT+E +N D + IAA +YAC + K + +
Sbjct 922 TSQPEQEVGTSEGKPISSLDAFPTQEGTLSPVNLTD-DQIAAGLYACTNNESTLKSIMK- 979
Query 77 AKKVTKKIINDAKK 90
KK K N KK
Sbjct 980 -KKDGNKDSNGTKK 992
> mcf:102141353 KANK1; KN motif and ankyrin repeat domains 1
Length=1364
Score = 34.7 bits (78), Expect = 1.9, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 38/74 (51%), Gaps = 3/74 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEADEYEKKLKRS 76
T+Q E+E GK + + + FPT+E +N D + IAA +YAC + K + +
Sbjct 922 TSQPEQEVGTSEGKPVNSLDAFPTQEDTLSPVNLTD-DQIAAGLYACTNNESTLKSIMK- 979
Query 77 AKKVTKKIINDAKK 90
KK K N AKK
Sbjct 980 -KKDGNKDSNGAKK 992
> mcc:697569 KANK1; KN motif and ankyrin repeat domains 1
Length=1194
Score = 34.7 bits (78), Expect = 1.9, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 38/74 (51%), Gaps = 3/74 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEADEYEKKLKRS 76
T+Q E+E GK + + + FPT+E +N D + IAA +YAC + K + +
Sbjct 764 TSQPEQEVGTSEGKPVNSLDAFPTQEDTLSPVNLTD-DQIAAGLYACTNNESTLKSIMK- 821
Query 77 AKKVTKKIINDAKK 90
KK K N AKK
Sbjct 822 -KKDGNKDSNGAKK 834
> cal:CaO19.9268 RKI1; similar to S. cerevisiae RKI1 (YOR095C)
pentose phosphate shunt enzyme ribose-5-phosphate ketol-isomerase
Length=263
Score = 34.3 bits (77), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 41/86 (48%), Gaps = 4/86 (5%)
Query 7 DMKDVFKVLPTTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEA 66
D KD F +PT + K+ +I G L T E++P + A + + VD L +I
Sbjct 72 DNKDSFICIPTGFQSKQLIIDNGLRLGTIEQYPDIDIAFDGADEVDPQL--NLIKGGGAC 129
Query 67 DEYEKKLKRSAKKVTKKIINDAKKKE 92
EK + SAKK ++ D +KK
Sbjct 130 LFQEKLVAASAKKFV--VVADYRKKS 153
> cal:CaO19.1701 RKI1; similar to S. cerevisiae RKI1 (YOR095C)
pentose phosphate shunt enzyme ribose-5-phosphate ketol-isomerase
Length=263
Score = 34.3 bits (77), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 41/86 (48%), Gaps = 4/86 (5%)
Query 7 DMKDVFKVLPTTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEA 66
D KD F +PT + K+ +I G L T E++P + A + + VD L +I
Sbjct 72 DNKDSFICIPTGFQSKQLIIDNGLRLGTIEQYPDIDIAFDGADEVDPQL--NLIKGGGAC 129
Query 67 DEYEKKLKRSAKKVTKKIINDAKKKE 92
EK + SAKK ++ D +KK
Sbjct 130 LFQEKLVAASAKKFV--VVADYRKKS 153
> lve:103081989 KANK1; KN motif and ankyrin repeat domains 1
Length=1351
Score = 34.3 bits (77), Expect = 2.4, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 39/74 (53%), Gaps = 3/74 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEADEYEKKLKRS 76
T++ E+E GK +++ + FPT+E+ +N D + IAA +Y C + K + +
Sbjct 920 TSRHEQEVGTTEGKPISSQDTFPTQESTPSPVNLTD-DQIAAGLYVCTNNESTLKSIMK- 977
Query 77 AKKVTKKIINDAKK 90
KK K + AKK
Sbjct 978 -KKDANKDSSGAKK 990
> ptg:102963812 KANK1; KN motif and ankyrin repeat domains 1
Length=1355
Score = 33.9 bits (76), Expect = 3.1, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 39/74 (53%), Gaps = 3/74 (4%)
Query 17 TTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEADEYEKKLKRS 76
T++ E+E G+ L + FP++E+A +N D + IAA +Y C ++ K + +
Sbjct 924 TSRHEQEAGTTDGRPLGGQDAFPSQESALSPVNLTD-DQIAAGLYVCTNSESTLKSIMK- 981
Query 77 AKKVTKKIINDAKK 90
KK + N AKK
Sbjct 982 -KKDGNRDSNGAKK 994
> gga:423043 SBF2; SET binding factor 2
Length=1845
Score = 33.1 bits (74), Expect = 5.7, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 24/41 (59%), Gaps = 0/41 (0%)
Query 50 SVDWNLIAAMIYACKEADEYEKKLKRSAKKVTKKIINDAKK 90
S DWN++AA A +EAD+ + + + +K+ +D KK
Sbjct 1589 SYDWNMVAAKCSALEEADQTDGSVSQKKRKIVWPCYDDVKK 1629
> cdu:CD36_81400 phosphoriboisomerase, putative (EC:5.3.1.6)
Length=263
Score = 32.3 bits (72), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 26/84 (31%), Positives = 41/84 (49%), Gaps = 4/84 (5%)
Query 9 KDVFKVLPTTQEEKEYMIVIGKHLATTEKFPTREAAEEKINSVDWNLIAAMIYACKEADE 68
KD F +PT + K+ +I G L T E++P + A + + VD +L +I
Sbjct 74 KDSFICIPTGFQSKQLIIDNGLKLGTIEQYPDIDIAFDGADEVDPHL--NLIKGGGACLF 131
Query 69 YEKKLKRSAKKVTKKIINDAKKKE 92
EK + SAKK ++ D +KK
Sbjct 132 QEKLVATSAKKFI--VVADYRKKS 153
Lambda K H a alpha
0.311 0.126 0.343 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 126744689040