bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-16_CDS_annotation_glimmer3.pl_2_3
Length=651
Score E
Sequences producing significant alignments: (Bits) Value
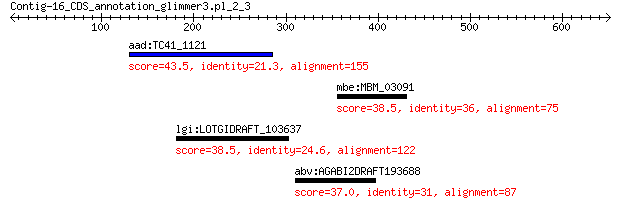
aad:TC41_1121 AMP-dependent synthetase and ligase 43.5 0.14
mbe:MBM_03091 hypothetical protein 38.5 5.2
lgi:LOTGIDRAFT_103637 hypothetical protein 38.5 5.6
abv:AGABI2DRAFT193688 AGABI2DRAFT_193688; hypothetical protein 37.0 8.9
> aad:TC41_1121 AMP-dependent synthetase and ligase
Length=408
Score = 43.5 bits (101), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 33/156 (21%), Positives = 67/156 (43%), Gaps = 20/156 (13%)
Query 130 EDEKQWSQINPSCLLSYLGIKGYGGMNNNATAPTATKNAVKVLGYWDIFKNFYANKQEEN 189
E +W + P L+ LGI G ++ A V +L +WD+ + ++E+
Sbjct 243 EPAAKWLVVTPIAHLTGLGIGCLGALSRGAP--------VALLSHWDVQRAVSVMRREQP 294
Query 190 YYMIGSNDPLSITINNTQITDPNNIPQNQGTVNAKSIIEIDDVLSIYNNSNVTLWVTQQI 249
Y++G+ P+ + + +++P +G V A + + + S+ N+ + +
Sbjct 295 TYLLGAT-PMLVDLARADDLTASSVPSLRGIVYAGATCPAEILRSL--NTKLGADIFACY 351
Query 250 GHSP-VKMTADEIGTTSTDNQKFIIQNLKIPTGNTW 284
G++ T+ +GT ST P+GN W
Sbjct 352 GYTEGASRTSRALGTVSTSP--------ACPSGNGW 379
> mbe:MBM_03091 hypothetical protein
Length=954
Score = 38.5 bits (88), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 27/85 (32%), Positives = 42/85 (49%), Gaps = 10/85 (12%)
Query 356 LKTYNSDLYQNWINTEWIDGANGINEITAVDV-SDGTLSMDALNLQQK----VYNMLNRI 410
+K YN L W TEWI IN +TA+ V S G LS+ A+ L++ +Y+ + ++
Sbjct 493 VKQYNDKLGARWFATEWISLDVAINGVTALGVQSIGPLSLQAMVLREPDLKLIYSRIAQL 552
Query 411 AVSGGS-----YRDWLETVYASGQY 430
G S Y +E+ G+Y
Sbjct 553 RAGGVSIGNSLYSQAVESFAKDGKY 577
> lgi:LOTGIDRAFT_103637 hypothetical protein
Length=580
Score = 38.5 bits (88), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 30/133 (23%), Positives = 60/133 (45%), Gaps = 11/133 (8%)
Query 181 FYANKQEENYYMIGSNDPLSITINNTQITDPNNIPQNQGTVNAKSIIEI----------D 230
F ++Q + Y + N + +ITD + P QG I+E+ +
Sbjct 151 FTMDRQRNSSYRVSRNSDIYRKSPAIKITDLDVSPDLQGLKIGDRILEVNGLNVRDKSVE 210
Query 231 DVLSIYNNSNVTLWVTQQIGHSPVKMTADEIGTTSTDN-QKFIIQNLKIPTGNTWWIRTI 289
D+ +I+NN+N L +T + SP++ ++D T T + + II N + ++
Sbjct 211 DIDTIFNNTNEVLHITLERDPSPLRHSSDSDSPTDTSSPDQVIIGNTPVQLRPRSSLKAR 270
Query 290 QSTQQQALVPFPL 302
+++++ P PL
Sbjct 271 NPSRRRSKSPSPL 283
> abv:AGABI2DRAFT193688 AGABI2DRAFT_193688; hypothetical protein
Length=216
Score = 37.0 bits (84), Expect = 8.9, Method: Composition-based stats.
Identities = 27/92 (29%), Positives = 44/92 (48%), Gaps = 15/92 (16%)
Query 310 DEILAKKGNQTFYLG----DGKSSTQIANV-FNERGDNQKLKTTRPQYGLLLKTYNSDLY 364
D + +G+Q Y+G DG+++T+ V ++ R D+++L+ P Y LLK +
Sbjct 32 DAVRKAQGSQHIYVGKCVDDGETNTRFVFVTWDSRADHKRLEEDAPVYEQLLKDF----- 86
Query 365 QNWINTEWIDGANGINEITAVDVSDGTLSMDA 396
T+ DGA G N V D T+ A
Sbjct 87 -----TKCADGAAGFNVFHIVSSDDPTMVFKA 113
Lambda K H a alpha
0.315 0.132 0.394 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1536646508365