bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-13_CDS_annotation_glimmer3.pl_2_1
Length=362
Score E
Sequences producing significant alignments: (Bits) Value
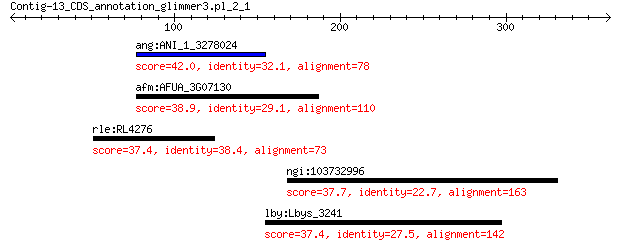
ang:ANI_1_3278024 An02g13590; two-component sensor protein his... 42.0 0.20
afm:AFUA_3G07130 sensor histidine kinase/response regulator 38.9 2.0
rle:RL4276 hypothetical protein 37.4 3.9
ngi:103732996 Myo3a; myosin IIIA 37.7 5.2
lby:Lbys_3241 alpha-l-fucosidase 37.4 5.6
> ang:ANI_1_3278024 An02g13590; two-component sensor protein histidine
protein kinase (dhkk, dhkj)
Length=1266
Score = 42.0 bits (97), Expect = 0.20, Method: Composition-based stats.
Identities = 25/79 (32%), Positives = 44/79 (56%), Gaps = 1/79 (1%)
Query 77 AMISRGLNPFVQGSAAMAGSRSPASGGAAASAASAPSLQAFRPDFSDVGSALA-SMAQAR 135
A +SRGL+ FV+GS++ + P+ G++A+A S P + R + VGS + M R
Sbjct 261 ARLSRGLSCFVEGSSSFVDASHPSLAGSSAAAPSTPPSSSQRANHLSVGSFNSFDMPSRR 320
Query 136 AAMINAEQNAALTPYKMQQ 154
+ +A ++++ YK Q
Sbjct 321 SHSTDARSFSSVSDYKADQ 339
> afm:AFUA_3G07130 sensor histidine kinase/response regulator
Length=1275
Score = 38.9 bits (89), Expect = 2.0, Method: Composition-based stats.
Identities = 32/111 (29%), Positives = 57/111 (51%), Gaps = 4/111 (4%)
Query 77 AMISRGLNPFVQGSAAMAGSRSPASGGAAASAASAPSLQAFRPDFSDVGSALA-SMAQAR 135
A +SRGL+ FV+GS++ S+ P+ G++A S P+ A R + GS+ + + R
Sbjct 290 ARLSRGLSYFVEGSSSFVDSQ-PSCSGSSAPTLSTPASSAMRANHLSAGSSHSFELPSKR 348
Query 136 AAMINAEQNAALTPYKMQQILGDTNYRNIGVGQSGYWTASTGRRSALLDQS 186
+ I+A +++++ K GD N + W A + R+ LD+S
Sbjct 349 SHSIDARSSSSVSDSKADN--GDQNNSSGPDTPLPDWWAGSRRKGLKLDES 397
> rle:RL4276 hypothetical protein
Length=265
Score = 37.4 bits (85), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 41/82 (50%), Gaps = 12/82 (15%)
Query 51 NAHQAQIQRDWQEKMWGMN-------NSYNSPNAMISRGLNPFVQGSAAMAGS--RSPAS 101
NA ++ QRD+ + G+ + NA +S P Q AA+ GS +PAS
Sbjct 135 NAVKSACQRDFMAQCSGVTPGGTEALSCLQQHNAALS---APCQQAVAALGGSAGSTPAS 191
Query 102 GGAAASAASAPSLQAFRPDFSD 123
GGAAA+ + P+ +A P FS
Sbjct 192 GGAAATVVTTPAPRAMMPAFSP 213
> ngi:103732996 Myo3a; myosin IIIA
Length=1612
Score = 37.7 bits (86), Expect = 5.2, Method: Composition-based stats.
Identities = 37/173 (21%), Positives = 70/173 (40%), Gaps = 18/173 (10%)
Query 168 QSGYWTASTGRRSALLDQSKEYQELKNMEFAGRLTSAQEAQILLDSEAQQVLNKYLDEQQ 227
+S S RR + KE LKN A T QE + + ++ ++ +Q
Sbjct 1083 ESAIIIQSAARRHLVRKTGKETANLKNTAVATIQTQDQETDYKKNFDTKR--ESFVKKQT 1140
Query 228 QADLFIKGQTLSNLYAQGSLTEAQYKTEMAKAVKTAAETNGIRINNKIASQTADSLIYAN 287
+ +F +S+ +GS + + T + KA TNG+ NN+ +Q S +Y
Sbjct 1141 ENAVFANESNISSPNNKGSPSIGKTSTALNKA------TNGVESNNRCQTQKKVSNVYRE 1194
Query 288 IQSNRARGLSSLWD--SKNTNVLKNIEYSK--------DKALRDYYKWSSKHK 330
+ W+ S +++E S+ D ++ YY+W ++ +
Sbjct 1195 EPHQELFIVGDPWEEVSSMQKYTQDLEESRKTGKHEKGDPVIQSYYQWYTEQR 1247
> lby:Lbys_3241 alpha-l-fucosidase
Length=793
Score = 37.4 bits (85), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 39/157 (25%), Positives = 74/157 (47%), Gaps = 30/157 (19%)
Query 155 ILGDTNYRNIGVGQSGYWTASTGRRSALLDQSKEYQELKNMEFAGRLTSAQEAQILLD-- 212
+ G T I + + W S G + A D+ + Y+ LK ++ Q+LL+
Sbjct 47 VFGQTAKERIALNEISLW--SGGPQDA--DREEAYKSLKPIQ-----------QLLLEGK 91
Query 213 -SEAQQVLNKYLDEQQQADLFIKG--------QTLSNLYAQ---GSLTEAQYKTEMAKAV 260
EAQ +L K + + F +G QTL +L+ + G ++ + ++ A+
Sbjct 92 NKEAQTLLEKEFIAKGRGSGFGRGAKDPYGSYQTLGDLFLEWKDGEVSNYKRWLDLDNAL 151
Query 261 KTAAET-NGIRINNKIASQTADSLIYANIQSNRARGL 296
T T NGI+I ++ + + LI+ ++S++A+GL
Sbjct 152 ATTQFTRNGIQITEEVFTDFKNDLIWVRLRSSKAKGL 188
Lambda K H a alpha
0.310 0.124 0.342 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 688208740258