bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-6_CDS_annotation_glimmer3.pl_2_9
Length=82
Score E
Sequences producing significant alignments: (Bits) Value
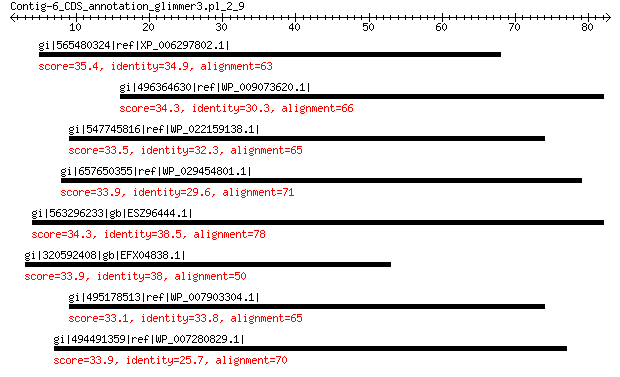
gi|565480324|ref|XP_006297802.1| hypothetical protein CARUB_v100... 35.4 2.7
gi|496364630|ref|WP_009073620.1| XRE family transcriptional regu... 34.3 4.9
gi|547745816|ref|WP_022159138.1| putative uncharacterized protein 33.5 5.0
gi|657650355|ref|WP_029454801.1| hypothetical protein 33.9 7.3
gi|563296233|gb|ESZ96444.1| potassium/sodium efflux P-type ATPase 34.3 7.8
gi|320592408|gb|EFX04838.1| two-component response regulator ssk1p 33.9 8.6
gi|495178513|ref|WP_007903304.1| hypothetical protein 33.1 8.9
gi|494491359|ref|WP_007280829.1| hypothetical protein 33.9 9.4
>gi|565480324|ref|XP_006297802.1| hypothetical protein CARUB_v10013838mg [Capsella rubella]
gi|482566511|gb|EOA30700.1| hypothetical protein CARUB_v10013838mg [Capsella rubella]
Length=409
Score = 35.4 bits (80), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 33/64 (52%), Gaps = 1/64 (2%)
Query 5 EIKEGETIETKVRRITEEKTPITDSAPIVYTNR-TDGVIAGYNIRTDRFEIALSAMDKVN 63
EIKE E++ I E+ T + S P+VY NR TDG +A + + E S D++
Sbjct 90 EIKESGIDESETVNIAEDVTQLIGSTPMVYLNRVTDGCLADIAAKLESMEPCRSVKDRIG 149
Query 64 KAKI 67
+ I
Sbjct 150 LSMI 153
>gi|496364630|ref|WP_009073620.1| XRE family transcriptional regulator [Metallosphaera yellowstonensis]
gi|373524622|gb|EHP69499.1| putative transcriptional regulator with C-terminal CBS domains
[Metallosphaera yellowstonensis MK1]
Length=174
Score = 34.3 bits (77), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 34/66 (52%), Gaps = 2/66 (3%)
Query 16 VRRITEEKTPITDSAPIVYTNRTDGVIAGYNIRTDRFEIALSAMDKVNKAKIATRNAKNK 75
VRRI EE TP+ +++ T VI Y +R + + M+K N +++ N+ K
Sbjct 38 VRRIMEELTPLIETSETASTIMHSPVIVAYP--DERIQEVVERMEKFNISQLPIVNSHGK 95
Query 76 VRGIRY 81
+ G+ Y
Sbjct 96 LVGMIY 101
>gi|547745816|ref|WP_022159138.1| putative uncharacterized protein [Prevotella sp. CAG:520]
gi|524349928|emb|CDB05206.1| putative uncharacterized protein [Prevotella sp. CAG:520]
Length=133
Score = 33.5 bits (75), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 32/65 (49%), Gaps = 2/65 (3%)
Query 9 GETIETKVRRITEEKTPITDSAPIVYTNRTDGVIAGYNIRTDRFEIALSAMDKVNKAKIA 68
E+IE + +R E T P++ RTD V+ N RT + S MD+++ AK+
Sbjct 19 HESIEKRAQREAREYTERNCPTPVLNYMRTDSVVFEMNTRT--YHYYCSIMDELDDAKVF 76
Query 69 TRNAK 73
N K
Sbjct 77 EMNRK 81
>gi|657650355|ref|WP_029454801.1| hypothetical protein [Candidatus Pelagibacter ubique]
Length=309
Score = 33.9 bits (76), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 21/78 (27%), Positives = 38/78 (49%), Gaps = 7/78 (9%)
Query 8 EGETIETKVRRITEEKTPITDSAPIVYTNRTDGVIAGYN--IRTD-----RFEIALSAMD 60
+ + +ET + E K P+ S V ++T+ V+ GYN + TD FE+++
Sbjct 155 QADIVETIIESTKESKLPVWLSISCVIDDQTNKVMLGYNDTLDTDTNVYEDFELSMDNFS 214
Query 61 KVNKAKIATRNAKNKVRG 78
K++ I ++ KV G
Sbjct 215 KIHNGPILIAHSDIKVTG 232
>gi|563296233|gb|ESZ96444.1| potassium/sodium efflux P-type ATPase [Sclerotinia borealis F-4157]
Length=1072
Score = 34.3 bits (77), Expect = 7.8, Method: Composition-based stats.
Identities = 30/110 (27%), Positives = 42/110 (38%), Gaps = 32/110 (29%)
Query 4 VEIKEGETIETKVR-------------------RITEEKTPITDS-------------AP 31
VE+K G+TI +R I +E TP+ DS +
Sbjct 157 VEVKTGDTIPGDIRLIETMNFETDEMILTGESLPIVKEATPVFDSDITAGDRINVAFSST 216
Query 32 IVYTNRTDGVIAGYNIRTDRFEIALSAMDKVNKAKIATRNAKNKVRGIRY 81
V R G++ +RT IA S +V+K + RNA K R RY
Sbjct 217 TVTKGRARGIVFAVGMRTQIGSIATSLRSEVSKIREVKRNADGKARPHRY 266
>gi|320592408|gb|EFX04838.1| two-component response regulator ssk1p [Grosmannia clavigera
kw1407]
Length=722
Score = 33.9 bits (76), Expect = 8.6, Method: Composition-based stats.
Identities = 19/56 (34%), Positives = 26/56 (46%), Gaps = 6/56 (11%)
Query 3 GVEIKEGETIETKVRRITEEKTPITD------SAPIVYTNRTDGVIAGYNIRTDRF 52
G I+E I+ +RRIT +P + P+ YT D +GY RTD F
Sbjct 87 GQTIEEALIIDVPLRRITPSSSPQAGRPDGLYAVPVGYTTSRDSYSSGYETRTDYF 142
>gi|495178513|ref|WP_007903304.1| hypothetical protein [Prevotella stercorea]
gi|357553825|gb|EHJ35563.1| hypothetical protein HMPREF0673_02961 [Prevotella stercorea DSM
18206]
Length=133
Score = 33.1 bits (74), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 22/65 (34%), Positives = 32/65 (49%), Gaps = 2/65 (3%)
Query 9 GETIETKVRRITEEKTPITDSAPIVYTNRTDGVIAGYNIRTDRFEIALSAMDKVNKAKIA 68
E+IE + +R E T P++ RTD V+ N RT + LS M+ ++ AKI
Sbjct 19 HESIEKRAQREAREYTEKNCPTPVLNYMRTDSVVFETNTRT--YHYYLSVMNDLDDAKIF 76
Query 69 TRNAK 73
N K
Sbjct 77 ELNRK 81
>gi|494491359|ref|WP_007280829.1| hypothetical protein [Lentisphaera araneosa]
gi|149136947|gb|EDM25373.1| hypothetical protein LNTAR_22045 [Lentisphaera araneosa HTCC2155]
Length=812
Score = 33.9 bits (76), Expect = 9.4, Method: Composition-based stats.
Identities = 18/71 (25%), Positives = 38/71 (54%), Gaps = 1/71 (1%)
Query 7 KEGETIETKVRRITEEKTPITDSAPIVYTNRTDGVIAGYN-IRTDRFEIALSAMDKVNKA 65
K+ +T V+ I +++ P+ P+ Y N D + + T+R A++ +++ K
Sbjct 245 KQNHQNDTLVKVIAQQELPVKQKIPVTYDNELDAYALDIDHLNTNRSPGAVNNREELIKV 304
Query 66 KIATRNAKNKV 76
+I RN+++KV
Sbjct 305 RIQNRNSEDKV 315
Lambda K H a alpha
0.314 0.131 0.348 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 434005500390