bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-53_CDS_annotation_glimmer3.pl_2_1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
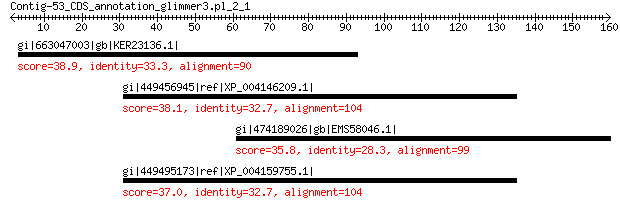
gi|663047003|gb|KER23136.1| hypothetical protein T265_08907 38.9 0.76
gi|449456945|ref|XP_004146209.1| PREDICTED: uncharacterized prot... 38.1 2.0
gi|474189026|gb|EMS58046.1| hypothetical protein TRIUR3_06689 35.8 4.5
gi|449495173|ref|XP_004159755.1| PREDICTED: uncharacterized LOC1... 37.0 4.7
>gi|663047003|gb|KER23136.1| hypothetical protein T265_08907 [Opisthorchis viverrini]
Length=232
Score = 38.9 bits (89), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 43/96 (45%), Gaps = 7/96 (7%)
Query 3 YTFCCRFSASIVIVPSSSGF---NAVIFEIPFSPSIQSVFTQLLKMSLSYVFSTSPPSLF 59
Y +C +S S VI P S VI E P SP I L LS + + P L
Sbjct 118 YAWCAGWS-SAVIQPDSRPIAHPRTVILESPTSPKIGKRLLSLKGHRLSVLMNLHPLCLK 176
Query 60 AD---AILIASNRLLTSLSPVSVIFNAIFPGCFNIS 92
+ ++ +A+ R LT++ P V+ I P C N+
Sbjct 177 QELELSVAVATFRCLTAIPPDGVMRAEILPSCSNLD 212
>gi|449456945|ref|XP_004146209.1| PREDICTED: uncharacterized protein LOC101212579 [Cucumis sativus]
Length=810
Score = 38.1 bits (87), Expect = 2.0, Method: Composition-based stats.
Identities = 34/107 (32%), Positives = 52/107 (49%), Gaps = 14/107 (13%)
Query 31 FSPSIQSVFTQLLKMSLSYVFS--TSPPSLFADAILIASNRLLTSLSPVSVIFNAIFPGC 88
FSPSI S+ L M L+Y+ S + PSL+ L+ +LL SL P F+A +
Sbjct 283 FSPSITSLDKSRLMMELNYMLSYGAAVPSLY----LLQRFKLLGSLLP----FHAAYLDK 334
Query 89 FNISSRILSRVSSFCSFKIVFS-EFLYNVSSPGNCPTWIPCIVVYPA 134
I S +SS K+ F+ + L + + P NC W+ + + A
Sbjct 335 QGIEK---SSLSSVMLMKLFFNLDKLVSCAHPSNCNIWVALLAFHLA 378
>gi|474189026|gb|EMS58046.1| hypothetical protein TRIUR3_06689 [Triticum urartu]
Length=130
Score = 35.8 bits (81), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 4/99 (4%)
Query 61 DAILIASNRLLTSLSPVSVIFNAIFPGCFNISSRILSRVSSFCSFKIVFSEFLYNVSSPG 120
DA +A+ L+ + +S + + GC NI +R+ + + C F V EF V S
Sbjct 32 DATRLAARNQLSQNTLLSGLVRDAYEGCLNI-NRVANEMLPNCRFAGVADEFARGVKSLE 90
Query 121 NCPTWIPCIVVYPASELPLENIVDTLSVVIALVVLFDVR 159
+C WI ++ P PL N+V + L+ L D +
Sbjct 91 SC--WIR-LMRPPVKSTPLYNLVWADRYKLLLIHLLDGK 126
>gi|449495173|ref|XP_004159755.1| PREDICTED: uncharacterized LOC101212579 [Cucumis sativus]
Length=647
Score = 37.0 bits (84), Expect = 4.7, Method: Composition-based stats.
Identities = 34/107 (32%), Positives = 52/107 (49%), Gaps = 14/107 (13%)
Query 31 FSPSIQSVFTQLLKMSLSYVFS--TSPPSLFADAILIASNRLLTSLSPVSVIFNAIFPGC 88
FSPSI S+ L M L+Y+ S + PSL+ L+ +LL SL P F+A +
Sbjct 207 FSPSITSLDKSRLMMELNYMLSYGAAVPSLY----LLQRFKLLGSLLP----FHAAYLDK 258
Query 89 FNISSRILSRVSSFCSFKIVFS-EFLYNVSSPGNCPTWIPCIVVYPA 134
I S +SS K+ F+ + L + + P NC W+ + + A
Sbjct 259 QGIEK---SSLSSVMLMKLFFNLDKLVSCAHPSNCNIWVALLAFHLA 302
Lambda K H a alpha
0.328 0.139 0.417 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 428605528881