bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_8
Length=294
Score E
Sequences producing significant alignments: (Bits) Value
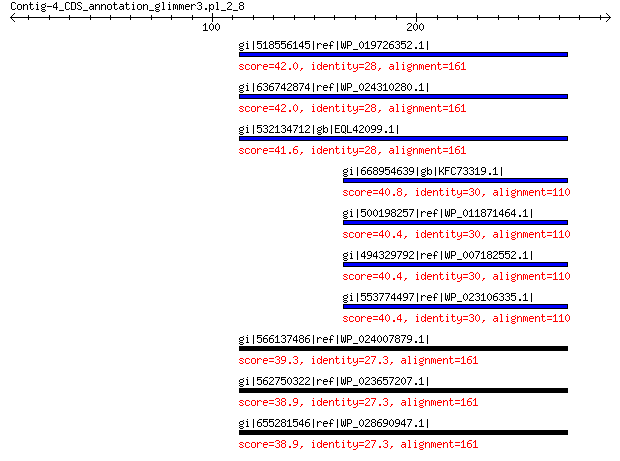
gi|518556145|ref|WP_019726352.1| MULTISPECIES: NADH dehydrogenase 42.0 0.34
gi|636742874|ref|WP_024310280.1| NADH dehydrogenase 42.0 0.40
gi|532134712|gb|EQL42099.1| hypothetical protein M770_06680 41.6 0.42
gi|668954639|gb|KFC73319.1| NADH dehydrogenase 40.8 1.0
gi|500198257|ref|WP_011871464.1| MULTISPECIES: NADH dehydrogenase 40.4 1.1
gi|494329792|ref|WP_007182552.1| NADH dehydrogenase 40.4 1.1
gi|553774497|ref|WP_023106335.1| hypothetical protein 40.4 1.4
gi|566137486|ref|WP_024007879.1| NADH dehydrogenase 39.3 3.1
gi|562750322|ref|WP_023657207.1| NADH dehydrogenase transmembran... 38.9 3.3
gi|655281546|ref|WP_028690947.1| MULTISPECIES: NADH dehydrogenase 38.9 3.4
>gi|518556145|ref|WP_019726352.1| MULTISPECIES: NADH dehydrogenase [Proteobacteria]
gi|542104497|gb|ERI33664.1| NADH dehydrogenase [Alcaligenes sp. EGD-AK7]
gi|552613155|gb|ERY29681.1| hypothetical protein Q074_02492 [Pseudomonas aeruginosa BL20]
Length=435
Score = 42.0 bits (97), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 45/163 (28%), Positives = 74/163 (45%), Gaps = 13/163 (8%)
Query 113 SNNLLVANIANSLLDADTKTILNKYLDQQQQAELNVK--AANYEYLVMSGQLKRQEVNNL 170
+N L A + +++D + L Q A L+ A YL + R EVN L
Sbjct 24 ANQLAGAEVDVTIIDRRNHHLFQPLLYQVAGASLSTSEIAWPIRYLFRN----RPEVNTL 79
Query 171 IAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGADAFHDSSI 230
+AE +E + + N LR+T D L+ AT T+ YFG + AF G D++
Sbjct 80 MAE-VEGIDQDARQVILNNGLRQTYDTLVLATGATHAYFG-HDEWGAFAPGLKTLEDATT 137
Query 231 LRSRAGSASESYKQSAFDTKLQPWREALNSTNMIFNGIGSGLD 273
+R R +A E ++++ P + A T +I G +G++
Sbjct 138 IRGRILAAFEEAERTS-----DPQQRAALQTFVIIGGGPTGVE 175
>gi|636742874|ref|WP_024310280.1| NADH dehydrogenase [Pseudomonas sp. P818]
Length=431
Score = 42.0 bits (97), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 45/163 (28%), Positives = 74/163 (45%), Gaps = 13/163 (8%)
Query 113 SNNLLVANIANSLLDADTKTILNKYLDQQQQAELNVK--AANYEYLVMSGQLKRQEVNNL 170
+N L A + +++D + L Q A L+ A YL + R EVN L
Sbjct 24 ANQLAGAEVDVTIIDRRNHHLFQPLLYQVAGASLSTSEIAWPIRYLFRN----RPEVNTL 79
Query 171 IAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGADAFHDSSI 230
+AE +E + + N LR+T D L+ AT T+ YFG + AF G D++
Sbjct 80 MAE-VEGIDQDARQVILNNGLRQTYDTLVLATGATHAYFG-HDEWGAFAPGLKTLEDATT 137
Query 231 LRSRAGSASESYKQSAFDTKLQPWREALNSTNMIFNGIGSGLD 273
+R R +A E ++++ P + A T +I G +G++
Sbjct 138 IRGRILAAFEEAERTS-----DPQQRAALQTFVIIGGGPTGVE 175
>gi|532134712|gb|EQL42099.1| hypothetical protein M770_06680 [Pseudomonas aeruginosa VRFPA03]
Length=337
Score = 41.6 bits (96), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 45/163 (28%), Positives = 74/163 (45%), Gaps = 13/163 (8%)
Query 113 SNNLLVANIANSLLDADTKTILNKYLDQQQQAELNVK--AANYEYLVMSGQLKRQEVNNL 170
+N L A + +++D + L Q A L+ A YL + R EVN L
Sbjct 24 ANQLAGAEVDVTIIDRRNHHLFQPLLYQVAGASLSTSEIAWPIRYLFRN----RPEVNTL 79
Query 171 IAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGADAFHDSSI 230
+AE +E + + N LR+T D L+ AT T+ YFG + AF G D++
Sbjct 80 MAE-VEGIDQDARQVILNNGLRQTYDTLVLATGATHAYFG-HDEWGAFAPGLKTLEDATT 137
Query 231 LRSRAGSASESYKQSAFDTKLQPWREALNSTNMIFNGIGSGLD 273
+R R +A E ++++ P + A T +I G +G++
Sbjct 138 IRGRILAAFEEAERTS-----DPQQRAALQTFVIIGGGPTGVE 175
>gi|668954639|gb|KFC73319.1| NADH dehydrogenase [Massilia sp. LC238]
Length=435
Score = 40.8 bits (94), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 7/110 (6%)
Query 164 RQEVNNLIAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGAD 223
R EVN L+AE +E + + N +R+T D L+ AT T+ YFG + AF G
Sbjct 73 RPEVNTLMAE-VEGIDQDARQVILNNGMRQTYDTLVLATGATHAYFG-HDEWGAFAPGLK 130
Query 224 AFHDSSILRSRAGSASESYKQSAFDTKLQPWREALNSTNMIFNGIGSGLD 273
D++ +R R +A E ++++ P + A T +I G +G++
Sbjct 131 TLEDATTIRGRILAAFEEAERTS-----DPQQRAALQTFVIIGGGPTGVE 175
>gi|500198257|ref|WP_011871464.1| MULTISPECIES: NADH dehydrogenase [Proteobacteria]
gi|134095227|ref|YP_001100302.1| NADH dehydrogenase transmembrane protein [Herminiimonas arsenicoxydans]
gi|528982810|ref|YP_008028127.1| NADH dehydrogenase [Achromobacter xylosoxidans NH44784-1996]
gi|133739130|emb|CAL62179.1| NADH dehydrogenase [Herminiimonas arsenicoxydans]
gi|507099418|emb|CCH05005.1| NADH dehydrogenase [Achromobacter xylosoxidans NH44784-1996]
gi|552502524|gb|ERX59559.1| hypothetical protein Q002_01730 [Pseudomonas aeruginosa CF18]
gi|574044158|gb|ETV20817.1| hypothetical protein Q050_00277 [Pseudomonas aeruginosa BWHPSA045]
Length=435
Score = 40.4 bits (93), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 7/110 (6%)
Query 164 RQEVNNLIAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGAD 223
R EVN L+AE +E + + N +R+T D L+ AT T+ YFG + AF G
Sbjct 73 RPEVNTLMAE-VEGIDQDARQVILNNGMRQTYDTLVLATGATHAYFG-HDEWGAFAPGLK 130
Query 224 AFHDSSILRSRAGSASESYKQSAFDTKLQPWREALNSTNMIFNGIGSGLD 273
D++ +R R +A E ++++ P + A T +I G +G++
Sbjct 131 TLEDATTIRGRILAAFEEAERTS-----DPQQRAALQTFVIIGGGPTGVE 175
>gi|494329792|ref|WP_007182552.1| NADH dehydrogenase [Burkholderia sp. Ch1-1]
gi|385182581|gb|EIF31857.1| NADH dehydrogenase, FAD-containing subunit [Burkholderia sp.
Ch1-1]
Length=435
Score = 40.4 bits (93), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 7/110 (6%)
Query 164 RQEVNNLIAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGAD 223
R EVN L+AE +E + + N +R+T D L+ AT T+ YFG + AF G
Sbjct 73 RPEVNTLMAE-VEGIDQDARQVILNNGMRQTYDTLVLATGATHAYFG-HDEWGAFAPGLK 130
Query 224 AFHDSSILRSRAGSASESYKQSAFDTKLQPWREALNSTNMIFNGIGSGLD 273
D++ +R R +A E ++++ P + A T +I G +G++
Sbjct 131 TLEDATTIRGRILAAFEEAERTS-----DPQQRAALQTFVIIGGGPTGVE 175
>gi|553774497|ref|WP_023106335.1| hypothetical protein [Pseudomonas aeruginosa]
gi|552322381|gb|ERV82540.1| hypothetical protein Q058_01628 [Pseudomonas aeruginosa BL04]
Length=435
Score = 40.4 bits (93), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 7/110 (6%)
Query 164 RQEVNNLIAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGAD 223
R EVN L+AE +E + + N +R+T D L+ AT T+ YFG + AF G
Sbjct 73 RPEVNTLMAE-VEGIDQDARQVILNNGMRQTYDTLVLATGATHAYFG-HDEWGAFAPGLK 130
Query 224 AFHDSSILRSRAGSASESYKQSAFDTKLQPWREALNSTNMIFNGIGSGLD 273
D++ +R R +A E ++++ P + A T +I G +G++
Sbjct 131 TLEDATTIRGRILAAFEEAERTS-----DPQQRAALQTFVIIGGGHTGVE 175
>gi|566137486|ref|WP_024007879.1| NADH dehydrogenase [Pseudomonas aeruginosa]
gi|564837582|gb|ETD49234.1| NADH dehydrogenase [Pseudomonas aeruginosa VRFPA08]
Length=435
Score = 39.3 bits (90), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 44/163 (27%), Positives = 73/163 (45%), Gaps = 13/163 (8%)
Query 113 SNNLLVANIANSLLDADTKTILNKYLDQQQQAELNVK--AANYEYLVMSGQLKRQEVNNL 170
+N L A + +++D + L Q A L+ A YL + R EVN L
Sbjct 24 ANELAGAEVDVTIIDRRNHHLFQPLLYQVAGASLSTSEIAWPIRYLFRN----RPEVNTL 79
Query 171 IAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGADAFHDSSI 230
+AE +E + + N R+T D L+ AT T+ YFG + AF G D++
Sbjct 80 MAE-VEGIDQDARQVILNNGSRQTYDTLVLATGATHAYFG-HDEWGAFAPGLKTLEDATT 137
Query 231 LRSRAGSASESYKQSAFDTKLQPWREALNSTNMIFNGIGSGLD 273
+R R +A E ++++ P + A T +I G +G++
Sbjct 138 IRGRILAAFEEAERTS-----DPQQRAALQTFVIIGGGPTGVE 175
>gi|562750322|ref|WP_023657207.1| NADH dehydrogenase transmembrane protein [Pseudomonas aeruginosa]
gi|560114867|emb|CDH76588.1| NADH dehydrogenase transmembrane protein [Pseudomonas aeruginosa
MH27]
gi|670354800|dbj|BAP22604.1| NADH dehydrogenase [Pseudomonas aeruginosa]
Length=435
Score = 38.9 bits (89), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 44/163 (27%), Positives = 73/163 (45%), Gaps = 13/163 (8%)
Query 113 SNNLLVANIANSLLDADTKTILNKYLDQQQQAELNVK--AANYEYLVMSGQLKRQEVNNL 170
+N L A + +++D + L Q A L+ A YL + R EVN L
Sbjct 24 ANQLAGAEVDVTIIDRRNHHLFQPLLYQVAGASLSTSEIAWPIRYLFRN----RPEVNTL 79
Query 171 IAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGADAFHDSSI 230
+AE +E + + N R+T D L+ AT T+ YFG + AF G D++
Sbjct 80 MAE-VEGIDQDARQVILNNGSRQTYDTLVLATGATHAYFG-HDEWGAFAPGLKTLEDATT 137
Query 231 LRSRAGSASESYKQSAFDTKLQPWREALNSTNMIFNGIGSGLD 273
+R R +A E ++++ P + A T +I G +G++
Sbjct 138 IRGRILAAFEEAERTS-----DPQQRAALQTFVIIGGGPTGVE 175
>gi|655281546|ref|WP_028690947.1| MULTISPECIES: NADH dehydrogenase [Pseudomonas putida group]
gi|633585547|gb|KDD56882.1| pyridine nucleotide-disulfide oxidoreductase [Bordetella bronchiseptica
OSU553]
gi|635747803|gb|KDF29411.1| hypothetical protein AE41_04907 [Enterobacter cloacae BIDMC 66]
Length=435
Score = 38.9 bits (89), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 44/163 (27%), Positives = 73/163 (45%), Gaps = 13/163 (8%)
Query 113 SNNLLVANIANSLLDADTKTILNKYLDQQQQAELNVK--AANYEYLVMSGQLKRQEVNNL 170
+N L A + +++D + L Q A L+ A YL + R EVN L
Sbjct 24 ANQLAGAEVDVTIIDRRNHHLFQPLLYQVAGASLSTSEIAWPIRYLFRN----RPEVNTL 79
Query 171 IAEEIETYARANGYNLQNRILRETSDGLIRATNNTNFYFGSYYHSRAFNAGADAFHDSSI 230
+AE +E + + N R+T D L+ AT T+ YFG + AF G D++
Sbjct 80 MAE-VEGIDQDARQVILNNGSRQTYDTLVLATGATHAYFG-HDEWGAFAPGLKTLEDATT 137
Query 231 LRSRAGSASESYKQSAFDTKLQPWREALNSTNMIFNGIGSGLD 273
+R R +A E ++++ P + A T +I G +G++
Sbjct 138 IRGRILAAFEEAERTS-----DPQQRAALQTFVIIGGGPTGVE 175
Lambda K H a alpha
0.313 0.127 0.358 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1540334286948