bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_7
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
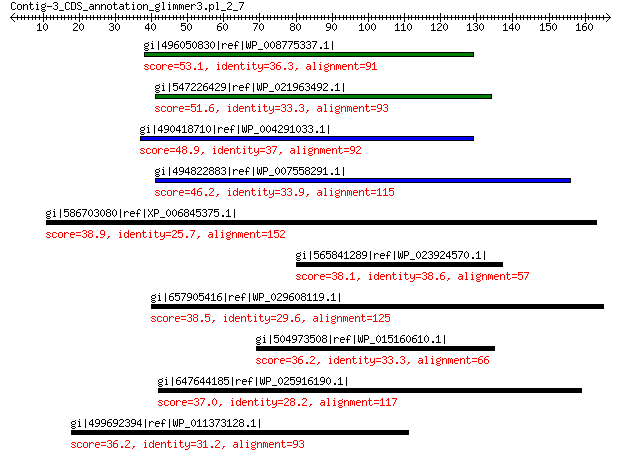
gi|496050830|ref|WP_008775337.1| hypothetical protein 53.1 3e-06
gi|547226429|ref|WP_021963492.1| putative uncharacterized protein 51.6 1e-05
gi|490418710|ref|WP_004291033.1| hypothetical protein 48.9 9e-05
gi|494822883|ref|WP_007558291.1| hypothetical protein 46.2 9e-04
gi|586703080|ref|XP_006845375.1| hypothetical protein AMTR_s0001... 38.9 0.85
gi|565841289|ref|WP_023924570.1| hypothetical protein 38.1 1.0
gi|657905416|ref|WP_029608119.1| ABC transporter substrate-bindi... 38.5 1.6
gi|504973508|ref|WP_015160610.1| hypothetical protein 36.2 2.0
gi|647644185|ref|WP_025916190.1| flagellar motor switch protein G 37.0 4.6
gi|499692394|ref|WP_011373128.1| resistance-nodulation-cell divi... 36.2 8.9
>gi|496050830|ref|WP_008775337.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448894|gb|EEO54685.1| hypothetical protein BSCG_01610 [Bacteroides sp. 2_2_4]
Length=154
Score = 53.1 bits (126), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/93 (35%), Positives = 51/93 (55%), Gaps = 7/93 (8%)
Query 38 EVDPVEQFRFETETFGESVSYRLRSDVSMLLHAADLAKRAGVSTVQRF--LDSKSPRSSS 95
E PV+QF F+ + S RL SD+ ML + L K + S ++ F + PR +
Sbjct 31 EESPVDQFLFQEVSVDGDTSIRLSSDIYMLFNQQRLDKLSQTSLLEYFNNISVTEPRFNE 90
Query 96 LQEQLDKLNPSDDELLSMVKSRHLQHPSEILAW 128
L+ +L D++L+S VKSR +Q SE++AW
Sbjct 91 LRSKL-----GDEQLISFVKSRFIQSKSELMAW 118
>gi|547226429|ref|WP_021963492.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103381|emb|CCY83992.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=152
Score = 51.6 bits (122), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/97 (32%), Positives = 54/97 (56%), Gaps = 7/97 (7%)
Query 41 PVEQFRFETETF----GESVSYRLRSDVSMLLHAADLAKRAGVSTVQRFLDSKSPRSSSL 96
P+ +F E T+ + + D+ ML + L G T+Q++LD +PRS SL
Sbjct 29 PIREFMTEKVTYFNGSDKKTAIAYVDDIYMLFNQNRL-DSVGRDTIQKWLDGLTPRSDSL 87
Query 97 QEQLDKLNPSDDELLSMVKSRHLQHPSEILAWIDSIN 133
+ + N +D++L+ + KSR++Q SE+LAW + +N
Sbjct 88 AKL--RENVTDEQLMEICKSRYIQSSSELLAWSEYLN 122
>gi|490418710|ref|WP_004291033.1| hypothetical protein [Bacteroides eggerthii]
gi|217986637|gb|EEC52971.1| hypothetical protein BACEGG_02722 [Bacteroides eggerthii DSM
20697]
Length=155
Score = 48.9 bits (115), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 34/95 (36%), Positives = 53/95 (56%), Gaps = 9/95 (9%)
Query 37 LEVDPVEQFRF-ETETFGESVSYRLRSDVSMLLHAADLAKRAGVSTVQRF--LDSKSPRS 93
+E P+ +F F E E G+ S R+ SD+ ML + L K V+ F L P+
Sbjct 31 VEPSPLHEFMFQEIECDGKK-SIRITSDIYMLFNQQRLDKLTRSQLVEYFDNLSVSEPKM 89
Query 94 SSLQEQLDKLNPSDDELLSMVKSRHLQHPSEILAW 128
S L++++ +D++L S VKSR +Q PSE++AW
Sbjct 90 SDLRKKM-----TDEQLCSFVKSRFIQTPSELMAW 119
>gi|494822883|ref|WP_007558291.1| hypothetical protein [Bacteroides plebeius]
gi|198272098|gb|EDY96367.1| hypothetical protein BACPLE_00803 [Bacteroides plebeius DSM 17135]
Length=140
Score = 46.2 bits (108), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 39/125 (31%), Positives = 67/125 (54%), Gaps = 16/125 (13%)
Query 41 PVEQF-RFETET-FGESVSYRLRSDVSMLLHAADLAKRAGVSTVQRFLD--SKSPRSSSL 96
PVE F R E +T G + + R+D+ M+L+ +R T+ +F D +S L
Sbjct 12 PVEDFYREEVQTPLGSTPAVTYRNDIYMILNQ----RRLDSMTLAQFSDYLDHDRSASQL 67
Query 97 QEQLDKLNPSDDELLSMVKSRHLQHPSEILAWIDSIN-ELAEDMRS-----EALKQTAEN 150
+ +K+ SD++L VKSR++QHPSE+ AW ++ E +++++ EA+K+ N
Sbjct 68 SQMREKM--SDEQLHQFVKSRYIQHPSELRAWASYLDIEYSKEIQKLEEAVEAVKKQTSN 125
Query 151 GSICP 155
P
Sbjct 126 TDPSP 130
>gi|586703080|ref|XP_006845375.1| hypothetical protein AMTR_s00019p00039340 [Amborella trichopoda]
gi|548847947|gb|ERN07050.1| hypothetical protein AMTR_s00019p00039340 [Amborella trichopoda]
Length=259
Score = 38.9 bits (89), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 39/153 (25%), Positives = 64/153 (42%), Gaps = 6/153 (4%)
Query 11 FGCGFREFTPTREISTFPSTRIGEISLEVDPVEQFRFETETFGESVSYRLRSDVSMLLHA 70
CG R P + +PST +G V +EQ R + V + D +L +A
Sbjct 76 LACGLRLLDP---LFVYPSTILGREKAIVVNLEQIR--CIITADEVLFLAFVDNYVLRYA 130
Query 71 ADLAKRAGVSTVQRFLDSKSPRSSSLQEQL-DKLNPSDDELLSMVKSRHLQHPSEILAWI 129
L KR + DS +P L L K P DEL S + + HL+ + + +
Sbjct 131 QLLQKRLTMKNDMGVGDSWAPVGGDLAAALVIKAYPLSDELTSKISTLHLEGVRRLKSRL 190
Query 130 DSINELAEDMRSEALKQTAENGSICPIVLVKKK 162
++ + +R E + ++G + + L KKK
Sbjct 191 VALTRRVQKVRDEIEQLMDDDGDMAEMYLTKKK 223
>gi|565841289|ref|WP_023924570.1| hypothetical protein [Prevotella nigrescens]
gi|564729908|gb|ETD29852.1| hypothetical protein HMPREF1173_00034 [Prevotella nigrescens
CC14M]
Length=148
Score = 38.1 bits (87), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 37/66 (56%), Gaps = 9/66 (14%)
Query 80 STVQRFLDSKSPRS------SSLQEQLDKLNPS---DDELLSMVKSRHLQHPSEILAWID 130
+ + R D +PR SSLQ+ + PS DD++LS++ R+ Q P EI ++D
Sbjct 56 TDLARLADPATPRKERDFILSSLQQLKGRSVPSELSDDDVLSLIPPRYCQDPVEIARFVD 115
Query 131 SINELA 136
++ +LA
Sbjct 116 AVKDLA 121
>gi|657905416|ref|WP_029608119.1| ABC transporter substrate-binding protein [Leucobacter chromiiresistens]
Length=490
Score = 38.5 bits (88), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 37/128 (29%), Positives = 62/128 (48%), Gaps = 13/128 (10%)
Query 40 DPVEQFRFETETFGESVSYRLRSDVSMLLHAADLAKRAGV-STVQRFL--DSKSPRSSSL 96
+P+ +E G + ++ LR DV+ H+ A V S+V+R DS+S R SS+
Sbjct 72 EPLVATGYEVSDDGLTYTFALRDDVA--FHSGKALTSADVKSSVERVTAPDSQSARKSSM 129
Query 97 QEQLDKLNPSDDELLSMVKSRHLQHPSEILAWIDSINELAEDMRSEALKQTAENGSICPI 156
+ D P D ++ +K R + P +L +I +N A D+ + AE+G+ P
Sbjct 130 EVVSDIATPDDQTVVFTLKQRSISFPY-LLTYIWIVNSEATDL------ENAEDGT-GPY 181
Query 157 VLVKKKNG 164
L + K G
Sbjct 182 TLGEWKRG 189
>gi|504973508|ref|WP_015160610.1| hypothetical protein [Chamaesiphon minutus]
gi|434387398|ref|YP_007098009.1| hypothetical protein Cha6605_3492 [Chamaesiphon minutus PCC 6605]
gi|428018388|gb|AFY94482.1| hypothetical protein Cha6605_3492 [Chamaesiphon minutus PCC 6605]
Length=95
Score = 36.2 bits (82), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 22/67 (33%), Positives = 36/67 (54%), Gaps = 1/67 (1%)
Query 69 HAADLAKRAGVSTVQRFLDSKSPRSSSLQEQLDKLNPSDDELLSMVKSRHLQHPSEI-LA 127
H DLA + TV++ ++ S ++Q+ D LNPS E+ + +S L PSE+ L
Sbjct 12 HILDLAIKHNDKTVKQLVNYPSSLLIAMQQYKDNLNPSYTEIYKIFESGLLLSPSEVDLN 71
Query 128 WIDSINE 134
W+ + N
Sbjct 72 WLKNQNN 78
>gi|647644185|ref|WP_025916190.1| flagellar motor switch protein G [Herminiimonas sp. CN]
Length=332
Score = 37.0 bits (84), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 33/124 (27%), Positives = 62/124 (50%), Gaps = 13/124 (10%)
Query 42 VEQFRFETETFGESVSYRLRSDVSMLLHAADLAKRAGVSTVQRFLDSKSPRSSSLQEQLD 101
+E+FRF+TE F S+S S + +L+ A + RA +Q L+S+ +S +D
Sbjct 55 MEEFRFDTEQFS-SISLDSDSYIRSVLNKALGSDRAA-DLIQEILESRDATTSG----MD 108
Query 102 KLNPSD-DELLSMVKSRHLQHPSEILAWIDSINE------LAEDMRSEALKQTAENGSIC 154
+LN + +E+ +++ H Q + +L +D + L E +R + + + A G +
Sbjct 109 RLNRLESNEVAELIRDEHPQIIATLLVHLDRLKAAEVLELLTERLRQDVILRVATFGGVQ 168
Query 155 PIVL 158
P L
Sbjct 169 PAAL 172
>gi|499692394|ref|WP_011373128.1| resistance-nodulation-cell division family transporter [Sulfurimonas
denitrificans]
gi|78777696|ref|YP_394011.1| resistance-nodulation-cell division family transporter [Sulfurimonas
denitrificans DSM 1251]
gi|78498236|gb|ABB44776.1| Resistance-Nodulation-Cell Division Superfamily transporter [Sulfurimonas
denitrificans DSM 1251]
Length=1005
Score = 36.2 bits (82), Expect = 8.9, Method: Composition-based stats.
Identities = 29/98 (30%), Positives = 48/98 (49%), Gaps = 6/98 (6%)
Query 18 FTPTREISTFPSTRIGEISLE---VDPVEQFRFETETFGESVSYRLRSDVSMLLHAADLA 74
F ++ +TF + EI E D ++ F + FGE S +L + V+ ++ D
Sbjct 728 FLEQKQSTTFNERGVMEIKTEDIKKDSIDTFLNFSIPFGEGKSVKL-TQVADIIEIRDYE 786
Query 75 K--RAGVSTVQRFLDSKSPRSSSLQEQLDKLNPSDDEL 110
K + S ++ F + R ++ QE LDKL P+ DEL
Sbjct 787 KINKLNGSIIKTFFATIDKRKTTSQEVLDKLEPTLDEL 824
Lambda K H a alpha
0.315 0.130 0.362 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 429014905530