bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-35_CDS_annotation_glimmer3.pl_2_5
Length=74
Score E
Sequences producing significant alignments: (Bits) Value
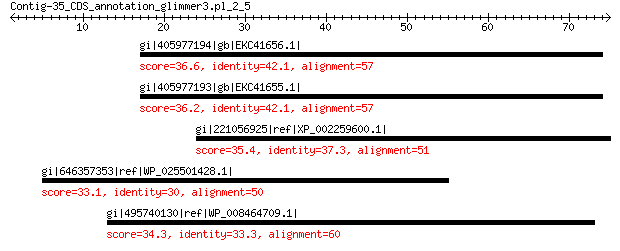
gi|405977194|gb|EKC41656.1| F-box only protein 8 36.6
gi|405977193|gb|EKC41655.1| Tctex1 domain-containing protein 1-B 36.2 0.89
gi|221056925|ref|XP_002259600.1| hypothetical protein, conserved... 35.4 2.8
gi|646357353|ref|WP_025501428.1| hypothetical protein 33.1 3.8
gi|495740130|ref|WP_008464709.1| collagen-binding protein 34.3 6.3
>gi|405977194|gb|EKC41656.1| F-box only protein 8 [Crassostrea gigas]
Length=434
Score = 36.6 bits (83), Expect = 0.87, Method: Composition-based stats.
Identities = 24/58 (41%), Positives = 34/58 (59%), Gaps = 2/58 (3%)
Query 17 GKTKKTRSMSGLTLLSGRQLTNTPPDKPEEESNPTNL-KDNNKNTKQKNPYRKDTTFL 73
G+ K +SG T L+GR+LT D PEE+S+ N K +NT + P +DTTF+
Sbjct 2 GRRKSIMKLSGTTALAGRRLTKALLDLPEEQSSIANKPKVRMENTYKMAP-DEDTTFI 58
>gi|405977193|gb|EKC41655.1| Tctex1 domain-containing protein 1-B [Crassostrea gigas]
Length=224
Score = 36.2 bits (82), Expect = 0.89, Method: Composition-based stats.
Identities = 24/58 (41%), Positives = 34/58 (59%), Gaps = 2/58 (3%)
Query 17 GKTKKTRSMSGLTLLSGRQLTNTPPDKPEEESNPTNL-KDNNKNTKQKNPYRKDTTFL 73
G+ K +SG T L+GR+LT D PEE+S+ N K +NT + P +DTTF+
Sbjct 66 GRRKSIMKLSGTTALAGRRLTKALLDLPEEQSSIANKPKVRMENTYKMAP-DEDTTFI 122
>gi|221056925|ref|XP_002259600.1| hypothetical protein, conserved in Plasmodium species [Plasmodium
knowlesi strain H]
gi|193809672|emb|CAQ40373.1| hypothetical protein, conserved in Plasmodium species [Plasmodium
knowlesi strain H]
Length=1515
Score = 35.4 bits (80), Expect = 2.8, Method: Composition-based stats.
Identities = 19/56 (34%), Positives = 30/56 (54%), Gaps = 5/56 (9%)
Query 24 SMSGLTLLSGRQLTNTPPDKPEEESNPTNL-----KDNNKNTKQKNPYRKDTTFLQ 74
S + + L+SG + N PP P +S+ KDNN+N ++KN + +FLQ
Sbjct 1250 SSTDVNLISGHKSDNMPPKNPPSDSDKIEQGSHSNKDNNQNQEEKNFINEMVSFLQ 1305
>gi|646357353|ref|WP_025501428.1| hypothetical protein [Vibrio parahaemolyticus]
gi|655688179|gb|KED80945.1| hypothetical protein EN02_07290 [Vibrio parahaemolyticus]
gi|655699294|gb|KED90871.1| hypothetical protein EM99_19995 [Vibrio parahaemolyticus]
gi|655707126|gb|KED97991.1| hypothetical protein EN03_02870 [Vibrio parahaemolyticus]
gi|655789041|gb|KEE70671.1| hypothetical protein EM83_22610 [Vibrio parahaemolyticus]
gi|655796923|gb|KEE77458.1| hypothetical protein EM49_12850 [Vibrio parahaemolyticus]
gi|655798347|gb|KEE78616.1| hypothetical protein EM65_12760 [Vibrio parahaemolyticus]
gi|655804693|gb|KEE83945.1| hypothetical protein EM78_03810 [Vibrio parahaemolyticus]
Length=90
Score = 33.1 bits (74), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 26/50 (52%), Gaps = 0/50 (0%)
Query 5 VLLYTRNGGSAQGKTKKTRSMSGLTLLSGRQLTNTPPDKPEEESNPTNLK 54
+ +YT N A+ + R T+ +G +L +TP KP E N T+++
Sbjct 37 IYIYTGNDAPAEFEGILVRDWISFTMKAGEKLFHTPAQKPHEAFNQTSIR 86
>gi|495740130|ref|WP_008464709.1| collagen-binding protein [Flavobacterium sp. F52]
gi|395436152|gb|EJG02091.1| TonB-dependent receptor, plug [Flavobacterium sp. F52]
Length=1080
Score = 34.3 bits (77), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 4/60 (7%)
Query 13 GSAQGKTKKTRSMSGLTLLSGRQLTNTPPDKPEEESNPTNLKDNNKNTKQKNPYRKDTTF 72
+A GK K S T+ G++ TN P + ++PT L D N NT +PY + F
Sbjct 937 ATASGKYKNLNS----TMADGKRWTNLDPATGQLVTDPTALADLNANTTMWSPYMRSAIF 992
Lambda K H a alpha
0.305 0.124 0.344 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 428645991228