bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-25_CDS_annotation_glimmer3.pl_2_1
Length=449
Score E
Sequences producing significant alignments: (Bits) Value
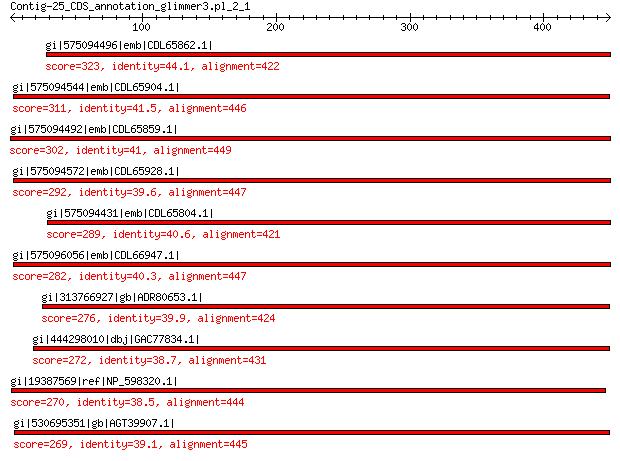
gi|575094496|emb|CDL65862.1| unnamed protein product 323 1e-100
gi|575094544|emb|CDL65904.1| unnamed protein product 311 6e-96
gi|575094492|emb|CDL65859.1| unnamed protein product 302 1e-92
gi|575094572|emb|CDL65928.1| unnamed protein product 292 1e-88
gi|575094431|emb|CDL65804.1| unnamed protein product 289 1e-87
gi|575096056|emb|CDL66947.1| unnamed protein product 282 1e-84
gi|313766927|gb|ADR80653.1| putative major coat protein 276 1e-82
gi|444298010|dbj|GAC77834.1| major capsid protein 272 8e-82
gi|19387569|ref|NP_598320.1| capsid protein 270 3e-80
gi|530695351|gb|AGT39907.1| major capsid protein 269 5e-80
>gi|575094496|emb|CDL65862.1| unnamed protein product [uncultured bacterium]
Length=568
Score = 323 bits (828), Expect = 1e-100, Method: Compositional matrix adjust.
Identities = 186/449 (41%), Positives = 256/449 (57%), Gaps = 31/449 (7%)
Query 28 GSIADYLGLPIGSISQSSPVSVLPFRCFALIYDKYFRNENTTDEIYIQKKGFSLSELIGA 87
G+IADY G+P G + S VS LPFR +ALI D++FR++N + I +L +
Sbjct 124 GTIADYFGIPTGVPNLS--VSALPFRAYALIVDEWFRDQNLQLPLNIPLDDTTLQGVNTG 181
Query 88 QNFSPNSYCGKLPKVNKYKDYFTSCVPNPQKGAPVTFNLGDQAVVRTSDSELVTGPQEQM 147
+ GK KY DYFTSC+P+PQKG VT V T D G + +
Sbjct 182 DYVTDTVKGGKPFVAAKYHDYFTSCLPSPQKGPDVTIAAVGDFPVYTGDPHNNNGSNKAL 241
Query 148 --ALTNSQSGSASVGEH----PLIVGLGGM-------RFDAAAFSGTVAAG--------- 185
++N SGS S + P ++ G + +A+ + T + G
Sbjct 242 HYGISNISSGSVSFSQGNYIIPSVLTTGSTQSVPAQGKLNASNITMTTSPGSPDSSFGSK 301
Query 186 --LYPNNLYADLSSANAISVDDLRLAFAYQKMLERDAIYGSRYNEYLYGHFGVHIPDAYI 243
+YP+NLYA SS A +++ LR+AF QK+ E+DA GSRY E + HF V DA +
Sbjct 302 LSVYPDNLYA--SSGTATTINQLRMAFQIQKLYEKDARAGSRYRELIRSHFSVTPLDARM 359
Query 244 QFPQYLGGGRTPLNIVQVAQTSQGTEESPLGNVGAYSWTNGRTG-YSRKFKEHGIVMTVA 302
Q P+YLGG R P+NI QV QTSQ ++ SP GNV S T+ G + + F EHG+++ VA
Sbjct 360 QVPEYLGGNRIPININQVVQTSQTSDVSPQGNVAGQSLTSDSHGDFIKSFTEHGMLIGVA 419
Query 303 CLRYRHTYQQGIAKKWRRKVREDFYDPLFSTIGQQPVYTTELYAQAFPQ--TVFGYREAW 360
RY HTYQQG++K W RK R D+Y P+ + IG+Q V E+YAQ Q VFGY+EAW
Sbjct 420 VARYDHTYQQGVSKLWSRKTRFDYYWPVLANIGEQAVLNKEIYAQGTAQDEEVFGYQEAW 479
Query 361 SELRNIPNTISGEMRSGVTNSLDIWHFADNYSSSPTLSQSFTEETPSYVDRTLSVPSSSQ 420
+E R P+ ++GEMRS SLD WHFAD+Y+S P LS + +E + +DR L+V SS
Sbjct 480 AEYRYKPSIVTGEMRSSARTSLDSWHFADDYNSLPKLSADWIKEDKTNIDRVLAVSSSVS 539
Query 421 NNFILNFYFDMSAVRKMPVYSMPSLIDHH 449
N + +FY + R +P YS+P LIDHH
Sbjct 540 NQYFADFYIENETTRALPFYSIPGLIDHH 568
>gi|575094544|emb|CDL65904.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 311 bits (796), Expect = 6e-96, Method: Compositional matrix adjust.
Identities = 185/462 (40%), Positives = 262/462 (57%), Gaps = 22/462 (5%)
Query 3 GNPNPSAYVNNVLEEIPMTYGEVNP---GSIADYLGLPIGSISQSSPVSVLPFRCFALIY 59
G SA++ ++P N G+IADY G+P G + V+ LPFR +ALI
Sbjct 95 GENTESAWLPTTEYQVPQVTAPANGWSIGTIADYFGIPTGV---ACSVNALPFRAYALIC 151
Query 60 DKYFRNENTTDEIYIQKKGFSLSELIGA--QNFSPNSYCGKLP-KVNKYKDYFTSCVPNP 116
+++FR+EN +D + I S + ++G+ N+ + G +P K KY DYFTSC+P P
Sbjct 152 NEWFRDENLSDPLNIP---ISDATVVGSNGDNYITDIVKGGMPFKACKYHDYFTSCLPAP 208
Query 117 QKGAPVTFNLGDQAVVRTSDSELVTGPQEQ---MALTNSQSGSASVGEHPL--IVGLGGM 171
QKG V L V T+ +V Q MA +S + S ++ + + G+ G
Sbjct 209 QKGPDVLLPLSSSPVPVTTSDTMVDPLQYSKYPMAGVDSWNLSPTLMRNIIRPFEGVEGA 268
Query 172 RFDAAAFSGTVAA--GLYPNNLYADLSSANAISVDDLRLAFAYQKMLERDAIYGSRYNEY 229
+ F+G + P NL A+L +A A S++ LRLAF Q++ ERDA G+RY E
Sbjct 269 NYQVHQFTGDIPTIDAFRPLNLVANLQNATAASINQLRLAFQIQRLYERDARGGTRYIEI 328
Query 230 LYGHFGVHIPDAYIQFPQYLGGGRTPLNIVQVAQTSQGTEESPLGN-VGAYSWTNGRTGY 288
L HFGV PDA +Q P+YLGG R P+NI QV Q S+ T SP GN VG T+ +
Sbjct 329 LKSHFGVTSPDARLQRPEYLGGNRIPININQVLQQSETTSTSPQGNPVGQSLTTDTNADF 388
Query 289 SRKFKEHGIVMTVACLRYRHTYQQGIAKKWRRKVREDFYDPLFSTIGQQPVYTTELYAQ- 347
+ F EHG V+ + RY HTYQQG+ + W RK R D+Y P+F+ IG+Q V E+Y
Sbjct 389 VKSFVEHGFVIGLMVARYDHTYQQGLERFWSRKDRFDYYWPVFAHIGEQAVLNKEIYTSG 448
Query 348 -AFPQTVFGYREAWSELRNIPNTISGEMRSGVTNSLDIWHFADNYSSSPTLSQSFTEETP 406
A VFGY+EA+++ R P+ ++GEMRS SLD+WH AD+Y+S P+LS S+ E+
Sbjct 449 TAVDDEVFGYQEAYADYRYKPSRVTGEMRSAAPQSLDVWHLADDYASLPSLSDSWIRESA 508
Query 407 SYVDRTLSVPSSSQNNFILNFYFDMSAVRKMPVYSMPSLIDH 448
S VDR L+V S+ + Y + R MP+YS+P LIDH
Sbjct 509 STVDRVLAVSSNVSAQLFCDIYIQNRSTRPMPMYSVPGLIDH 550
>gi|575094492|emb|CDL65859.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 302 bits (773), Expect = 1e-92, Method: Compositional matrix adjust.
Identities = 184/466 (39%), Positives = 258/466 (55%), Gaps = 25/466 (5%)
Query 1 VFGNPNPSAYVNNVLEEIPMTY---GEVNPGSIADYLGLPIGSISQSSPVSVLPFRCFAL 57
+ G SA+ V +P G N G+IADY+G+P G S V+ +PFR +AL
Sbjct 94 LMGENTQSAWTPQVEYSVPQITAPEGGWNVGTIADYMGIPTGVSGLS--VNAMPFRAYAL 151
Query 58 IYDKYFRNENTTDEIYIQKKGFSLSELIGAQNFSPNSYCGKLP-KVNKYKDYFTSCVPNP 116
I +++FR+EN TD + I G + + + + G LP K KY DYFTSC+P P
Sbjct 152 ICNEWFRDENLTDPLNI-PVGDATVAGVNTGTYVTDVAKGGLPFKAAKYHDYFTSCLPAP 210
Query 117 QKGAPVTFNLGDQAVVRTS------DSELVTGPQEQMALTNSQSGSASVGEHPLIVGLGG 170
QKG V + +V + DS V P + + S SV G G
Sbjct 211 QKGPDVLISAVGSGIVPVTATDNDNDSLNVNSPGMRFV----GNSSTSVNYLAFGGGDGY 266
Query 171 MRFDAAAFSGTV-AAGLYPNNLYADLSSANAI---SVDDLRLAFAYQKMLERDAIYGSRY 226
+ D S + + P NL+ADLS+A + +++ LR AF QK+ ERDA G+RY
Sbjct 267 VVTDTPKPSTPIHGISMIPTNLWADLSTATDLPVATINQLRTAFQIQKLYERDARGGTRY 326
Query 227 NEYLYGHFGVHIPDAYIQFPQYLGGGRTPLNIVQVAQTSQGTEESPLGNVGAYSWT-NGR 285
E L HFGV PDA +Q P+YLGG R P+NI QV Q+S+ T +P GN AYS T +
Sbjct 327 IEILKSHFGVTSPDARLQRPEYLGGSRVPININQVIQSSE-TGATPQGNAAAYSLTTDSH 385
Query 286 TGYSRKFKEHGIVMTVACLRYRHTYQQGIAKKWRRKVREDFYDPLFSTIGQQPVYTTELY 345
+ +++ F EHG ++ + RY H+YQQG+ + W RK R D+Y P+F+ +G+ V E++
Sbjct 386 SEFTKSFVEHGFIIGLMVARYDHSYQQGLQRFWSRKDRFDYYWPVFANLGEMAVKNKEIF 445
Query 346 AQA--FPQTVFGYREAWSELRNIPNTISGEMRSGVTNSLDIWHFADNYSSSPTLSQSFTE 403
AQ VFGY+EAW++ R P+ ++GEMRS SLDIWH AD+Y + P+LS S+
Sbjct 446 AQGTDVDDEVFGYQEAWADYRYKPSVVTGEMRSQYAQSLDIWHLADDYENLPSLSDSWIR 505
Query 404 ETPSYVDRTLSVPSSSQNNFILNFYFDMSAVRKMPVYSMPSLIDHH 449
E S V+R L+V S + Y A R MP+YS+P LIDHH
Sbjct 506 EDSSTVNRVLAVSDSVSAQLFCDIYIRCLATRPMPLYSIPGLIDHH 551
>gi|575094572|emb|CDL65928.1| unnamed protein product [uncultured bacterium]
Length=556
Score = 292 bits (747), Expect = 1e-88, Method: Compositional matrix adjust.
Identities = 177/468 (38%), Positives = 252/468 (54%), Gaps = 28/468 (6%)
Query 3 GNPNPSAYVNNVLEEIPMTY---GEVNPGSIADYLGLPIGSISQSSPVSVLPFRCFALIY 59
G SA++ V +IP G N G++ADY G+P G S V+ LPFR +AL+
Sbjct 96 GENTQSAWIPEVEYQIPQLTAPEGGWNIGTLADYFGIPTGVSGIS--VNALPFRAYALVC 153
Query 60 DKYFRNENTTDEIYIQKKGFSLSELIGAQNFSPNSYCGKLP-KVNKYKDYFTSCVPNPQK 118
+++FR++N +D + I +++ + F + G LP KY DYFTSC+P PQK
Sbjct 154 NEWFRDQNLSDPLNIPVGDATVTG-VNTGTFITDVVKGGLPYTAAKYHDYFTSCLPAPQK 212
Query 119 GAPVTF------NLGDQAVVRTSDSELVTGPQEQ--MALTNSQSGSASVGEHPLIVGLGG 170
G VT NL + T D+ GP + + + NS+ +
Sbjct 213 GPDVTIPVTSGHNLPVMFLNETHDA----GPYKPFGVGIQNSELRNFYGFGSGSSGATST 268
Query 171 MRFDAAAFSGTVAAGL-----YPNNLYA-DLSSANAISVDDLRLAFAYQKMLERDAIYGS 224
+ G+ G+ P N++A + +++ LRLAF QK+ E+DA G+
Sbjct 269 SDTSSTVEVGSDGTGIGQNFWTPTNMWAVESGDVGMATINQLRLAFQLQKLYEKDARGGT 328
Query 225 RYNEYLYGHFGVHIPDAYIQFPQYLGGGRTPLNIVQVAQTSQGTEESPLGNVGAYSWTNG 284
RY E + HFGV PD+ +Q P+YLGG R P+N+ Q+ Q SQ TE+SPLG + S T
Sbjct 329 RYTEIIRSHFGVVSPDSRLQRPEYLGGNRIPINVNQIIQQSQSTEQSPLGALAGMSVTTD 388
Query 285 R-TGYSRKFKEHGIVMTVACLRYRHTYQQGIAKKWRRKVREDFYDPLFSTIGQQPVYTTE 343
+ + + + F EHG ++ + RY HTYQQG+ + W RK R DFY P+ + IG+Q V E
Sbjct 389 KNSDFIKSFVEHGYIIGLVVARYDHTYQQGLDRMWSRKDRFDFYWPVLANIGEQAVLNKE 448
Query 344 LYAQA--FPQTVFGYREAWSELRNIPNTISGEMRSGVTNSLDIWHFADNYSSSPTLSQSF 401
+Y VFGY+EAW+E R PN + GEMRS SLD+WH D+YSS P LS S+
Sbjct 449 IYIDGSDTDDEVFGYQEAWAEYRYKPNRVCGEMRSSAPQSLDVWHLGDDYSSLPYLSDSW 508
Query 402 TEETPSYVDRTLSVPSSSQNNFILNFYFDMSAVRKMPVYSMPSLIDHH 449
E + VDR L+V SS + + Y A R MP+YS+P LIDHH
Sbjct 509 IREDKTNVDRVLAVTSSVSDQLFADIYICNKATRPMPMYSIPGLIDHH 556
>gi|575094431|emb|CDL65804.1| unnamed protein product [uncultured bacterium]
Length=560
Score = 289 bits (740), Expect = 1e-87, Method: Compositional matrix adjust.
Identities = 171/444 (39%), Positives = 252/444 (57%), Gaps = 30/444 (7%)
Query 29 SIADYLGLP--IGSISQSSPVSVLPFRCFALIYDKYFRNENTTDEIYIQKKGFSLSELIG 86
S+AD++G+P + +IS V+ LPFR + LIY+++FRN+N T+ ++ +++
Sbjct 124 SLADHMGIPTKVDNIS----VNALPFRAYGLIYNEFFRNQNLTNPTQVEVTDANIAGKNP 179
Query 87 AQNFSPNSYC---GKLPKVNKYKDYFTSCVPNPQKGAPVTFNL---------GD--QAVV 132
+ N + K K K+ DYFT +P PQKG PV NL GD +
Sbjct 180 NDVKNSNDWAITGAKCLKSAKFFDYFTGALPQPQKGEPVEINLASSWLPVGIGDYHGPLD 239
Query 133 RTSDSELVTGPQEQMALTNSQSGSASVGEHPLIVGLGGMR-FDAAA---FSGTVAAGLYP 188
+ S+S+ +T ++ + + + V G++ F+ A FS + A YP
Sbjct 240 KVSNSDTLTWESPSSEGNTKRTYALGMVQQEGEVNPNGLKNFETKAGGSFSESGAVAAYP 299
Query 189 NNLYADLSSANAISVDDLRLAFAYQKMLERDAIYGSRYNEYLYGHFGVHIPDAYIQFPQY 248
NL+A +A A +V+ LR AF QK+LE+DA G+RY E L HFGV DA +Q P+Y
Sbjct 300 TNLWASPVTA-AATVNQLRQAFQVQKLLEKDARGGTRYREILKNHFGVTTSDARMQIPEY 358
Query 249 LGGGRTPLNIVQVAQTSQGTEESPLGNVGAYSWTN-GRTGYSRKFKEHGIVMTVACLRYR 307
LGG + P+N+ QV QTS T+ SP GN A S T ++ +++ F EHG ++ VA R
Sbjct 359 LGGCKVPINVSQVVQTSASTDASPQGNTAAISVTPFSKSMFTKSFDEHGFIIGVATARTA 418
Query 308 HTYQQGIAKKWRRKVREDFYDPLFSTIGQQPVYTTELYAQ--AFPQTVFGYREAWSELRN 365
+YQQGI + W RK R D+Y P+ + IG+Q + E+YAQ A FGY+EAW++ R
Sbjct 419 QSYQQGIERMWSRKDRLDYYFPVLANIGEQAILNKEIYAQGNAKDDEAFGYQEAWADYRY 478
Query 366 IPNTISGEMRSGVTNSLDIWHFADNYSSSPTLSQSFTEETPSYVDRTLSVPSSSQNNFIL 425
PNTI G RS SLD WH+ +Y PTLS + E++ + RTL+V ++ +FI
Sbjct 479 KPNTICGRFRSNAQQSLDAWHYGQDYDKLPTLSTDWMEQSDIEMKRTLAV--QTEPDFIA 536
Query 426 NFYFDMSAVRKMPVYSMPSLIDHH 449
NF F+ VR MP+YS+P LIDH+
Sbjct 537 NFRFNCKTVRVMPLYSIPGLIDHN 560
>gi|575096056|emb|CDL66947.1| unnamed protein product [uncultured bacterium]
Length=570
Score = 282 bits (721), Expect = 1e-84, Method: Compositional matrix adjust.
Identities = 180/479 (38%), Positives = 247/479 (52%), Gaps = 36/479 (8%)
Query 3 GNPNPSAYVNNVLEEIPMT---YGEVNPGSIADYLGLPIGSISQSSPVSVLPFRCFALIY 59
G N SA++ IP G G+IADY GLP G + S VS LPFR +ALI
Sbjct 96 GENNESAWIPQTEYAIPQLKSPVGGFEVGTIADYFGLPTGVANLS--VSALPFRAYALIM 153
Query 60 DKYFRNENTTDEIYIQKKGFSLSELIGAQNFSPNSYCGKLPKVNKYKDYFTSCVPNPQKG 119
+++FR+EN D + + +++ + + + GK KY DYFTS +P PQKG
Sbjct 154 NEWFRDENLMDPLVVPTDDATVTGVNTGIFVTDVAKGGKPFVAAKYHDYFTSALPAPQKG 213
Query 120 APVTF---NLGDQAVVRTSDSELVTGPQEQMALTNSQSGSASVGEHPLIVGL-------- 168
V + G+ VV ++ + + N SGS G G+
Sbjct 214 PDVVIPVASAGNYNVVGNGKGLALSDGSKMSIICNGLSGSNGQGTELFASGILGSQVGSS 273
Query 169 ------GGMRFDAAAFSGTVAAGLYPNNL------YADLSSANAISVDDLRLAFAYQKML 216
+R D AA L NNL +A A +++ LR+AF QK
Sbjct 274 GGFGSGSSLRGDGIILGVPTAAQL-GNNLENSGLIAIASGNAAAATINQLRMAFQIQKFY 332
Query 217 ERDAIYGSRYNEYLYGHFGVHIPDAYIQFPQYLGGGRTPLNIVQVAQTSQGT---EESPL 273
E+ A GSRY E + FGV PDA +Q +YLGG R P+NI QV Q S GT +P
Sbjct 333 EKQARGGSRYTEVIRSFFGVTSPDARLQRSEYLGGNRIPININQVIQQS-GTGSASTTPQ 391
Query 274 GNV-GAYSWTNGRTGYSRKFKEHGIVMTVACLRYRHTYQQGIAKKWRRKVREDFYDPLFS 332
G V G T+ + +++ F EHG ++ V C RY HTYQQGI + W RK + D+Y P+FS
Sbjct 392 GTVVGMSQTTDTHSDFTKSFTEHGFIIGVMCARYDHTYQQGIDRMWSRKDKFDYYWPVFS 451
Query 333 TIGQQPVYTTELYAQ--AFPQTVFGYREAWSELRNIPNTISGEMRSGVTNSLDIWHFADN 390
IG+Q + E+YAQ A VFGY+EAW+E R P+ ++GEMRS SLD+WH AD+
Sbjct 452 NIGEQAIKNKEIYAQGNATDDEVFGYQEAWAEYRYKPSRVTGEMRSSYAQSLDVWHLADD 511
Query 391 YSSSPTLSQSFTEETPSYVDRTLSVPSSSQNNFILNFYFDMSAVRKMPVYSMPSLIDHH 449
YS P+LS + E ++R L+V + N F + Y R MP+YS+P LIDHH
Sbjct 512 YSKLPSLSDEWIREDAKTLNRVLAVSDQNSNQFFADIYVKNLCTRPMPMYSIPGLIDHH 570
>gi|313766927|gb|ADR80653.1| putative major coat protein [Uncultured Microviridae]
Length=533
Score = 276 bits (705), Expect = 1e-82, Method: Compositional matrix adjust.
Identities = 169/430 (39%), Positives = 240/430 (56%), Gaps = 42/430 (10%)
Query 25 VNPGSIADYLGLP--IGSISQSSPVSVLPFRCFALIYDKYFRNENTTDEIYIQKKGFSLS 82
V GS+ DY+GLP I I ++ L R + LI++++FR+EN D + + K
Sbjct 124 VAEGSLFDYMGLPTQIAGIDFNN----LHGRAYNLIWNEWFRDENLQDSLGVPKDD---- 175
Query 83 ELIGAQNFSPNSYCG-KLPKVNKYKDYFTSCVPNPQKGAPVTFNLGDQAVVRTSDSELVT 141
P++Y G + K K DYFTS +P PQKG V+ LG A + T+
Sbjct 176 --------GPDTYTGYTIQKRGKRHDYFTSALPWPQKGDAVSLPLGTSADIHTA------ 221
Query 142 GPQEQMALTNSQSGSASVGEHPLIVGLGGMRFDAAAFSGTVAAGLYPNNLYADLSSANAI 201
A + G SVG + L + A GT N ++ADLS+A A
Sbjct 222 ------AAAGTDIGIYSVGSSDFRL-LTSDPVEVALSGGTPPE---TNKMFADLSNATAA 271
Query 202 SVDDLRLAFAYQKMLERDAIYGSRYNEYLYGHFGVHIPDAYIQFPQYLGGGRTPLNIVQV 261
+++ LR AF Q++ E+DA G+RY E L HFGV PDA +Q P+YLGG +T + + V
Sbjct 272 TINQLREAFQIQRLYEKDARGGTRYTEILQSHFGVTSPDARLQRPEYLGGQKTEVMMQTV 331
Query 262 AQTSQGTEESPLGNVGAYSWTNGRTGYSRKFKEHGIVMTVACLRYRHTYQQGIAKKWRRK 321
QTS SP GN+ A R G+S+ F EHG+++ +AC+ TYQQG+ + W R+
Sbjct 332 PQTSSTDSTSPQGNLAALGTATSRGGFSKSFVEHGVLIGLACVFADLTYQQGMNRMWSRR 391
Query 322 VREDFYDPLFSTIGQQPVYTTELYAQ---AFPQTVFGYREAWSELRNIPNTISGEMRSGV 378
R DFY P + +G+Q V E+Y Q A QT FGY+E ++E R P+ I+G+MRS
Sbjct 392 DRWDFYWPSLAHLGEQAVLNQEIYTQGTSADTQT-FGYQERFAEYRYKPSQITGKMRSNA 450
Query 379 TNSLDIWHFADNYSSSPTLSQSFTEETPSYVDRTLSVPSSSQNNFILNFYFDMSAVRKMP 438
T +LD WH A ++++ P L+ SF EE P VDR ++VPS + FI ++YFD+ R MP
Sbjct 451 TGTLDAWHLAQDFTALPALNASFIEENPP-VDRVIAVPSEPE--FIWDWYFDLKTTRPMP 507
Query 439 VYSMPSLIDH 448
VYS+P LIDH
Sbjct 508 VYSVPGLIDH 517
>gi|444298010|dbj|GAC77834.1| major capsid protein [uncultured marine virus]
Length=480
Score = 272 bits (695), Expect = 8e-82, Method: Compositional matrix adjust.
Identities = 167/441 (38%), Positives = 245/441 (56%), Gaps = 36/441 (8%)
Query 18 IPMTYGEVNPGSIADYLGLPIGSISQSSPVSVLPFRCFALIYDKYFRNENTTDEIYIQKK 77
IP V GS+AD++GLP+G ++ LPFRC+ LIY+++FR+EN D I
Sbjct 65 IPKMVAAVIEGSLADHMGLPLGG---QFSINALPFRCYNLIYNEWFRDENLQDSIPENTD 121
Query 78 GFSLSELIGAQNFSPNSYCG-KLPKVNKYKDYFTSCVPNPQKGAPVTFNLGDQA-VVRTS 135
P+S L + K DYFTSC+P PQKGA VT LG A +
Sbjct 122 D------------GPDSVLDYLLRRRGKRHDYFTSCLPWPQKGAAVTLPLGTSAPITGIG 169
Query 136 DSELVTGPQEQMALTNSQSGSASVGEHPLIVGLGGMRFDAAAFSGTVAAGLYPNNLYADL 195
D V Q + T+ +G+ S H I F AF +PN + ADL
Sbjct 170 DQLQVYAGQTDVHETDG-TGTVSYANH--IESATAASF---AFEEDPDNAGFPN-IRADL 222
Query 196 SSANAISVDDLRLAFAYQKMLERDAIYGSRYNEYLYGHFGVHIPDAYIQFPQYLGGGRTP 255
++A A +++ LR AF QK+LERDA G+RY E + HF V PD+ +Q P+YLGGG +
Sbjct 223 TNATAATINQLRQAFQIQKLLERDARGGTRYTEIIRAHFSVLSPDSRLQRPEYLGGGSSN 282
Query 256 LNIVQVAQTSQGT----EESPLGNVGAY--SWTNGRTGYSRKFKEHGIVMTVACLRYRHT 309
+NI +AQT + +E+P GN+ A S +G G+++ F EHG ++ + +R T
Sbjct 283 INITPIAQTQRSDTTTPDETPQGNLAAIGTSAFSGH-GFTKSFTEHGYILGLCEVRADLT 341
Query 310 YQQGIAKKWRRKVREDFYDPLFSTIGQQPVYTTELYAQAFP--QTVFGYREAWSELRNIP 367
YQQGI + W R R DFY P S IG+Q V + E++A A + VFGY+E ++E R P
Sbjct 342 YQQGIDRLWSRDTRYDFYWPALSHIGEQAVLSKEIFADATAGDEDVFGYQERFAEYRYKP 401
Query 368 NTISGEMRSGVTNSLDIWHFADNYSSSPTLSQSFTEETPSYVDRTLSVPSSSQNNFILNF 427
+ IS RS SLD+WH + ++++ P L+ +F E+TP +DR ++V + + + +L+
Sbjct 402 SRISSLFRSNAAASLDVWHLSQDFAARPVLNSTFIEDTPP-IDRVIAV--TDEPHILLDA 458
Query 428 YFDMSAVRKMPVYSMPSLIDH 448
YF + R +P+Y +P LIDH
Sbjct 459 YFKLRCARPLPLYGVPGLIDH 479
>gi|19387569|ref|NP_598320.1| capsid protein [Spiroplasma phage 4]
gi|137968|sp|P11333.1|F_SPV4 RecName: Full=Capsid protein VP1; AltName: Full=VP1 [Spiroplasma
phage 4]
gi|334999|gb|AAA72621.1| capsid protein [Spiroplasma phage 4]
Length=553
Score = 270 bits (689), Expect = 3e-80, Method: Compositional matrix adjust.
Identities = 171/458 (37%), Positives = 248/458 (54%), Gaps = 21/458 (5%)
Query 2 FGNPNPSAYVNNV--LEEIPMTYGEVNPGSIADYLGLPIGSISQSSP---VSVLPFRCFA 56
FG + S V N + +I G + G++AD+ G I+ P V L FR +A
Sbjct 100 FGENSDSWDVKNAPPVPDIVAPSGGWDYGTLADHFG-----ITPKVPGIRVKSLRFRAYA 154
Query 57 LIYDKYFRNENTTDEIYIQKKGFSLSELIGAQNFSPNSYCGKLPKVNKYKDYFTSCVPNP 116
I + +FR++N + E + + G+ + GK NKY DYFTSC+P P
Sbjct 155 KIINDWFRDQNLSSECALTLDSSNSQGSNGSNQVTDIQLGGKPYIANKYHDYFTSCLPAP 214
Query 117 QKGAPVTFNLGDQAVVRTSDSELVTGPQEQMALTNSQSGSASVGEHPLIVGLGGMRFDAA 176
QKGAP T N+G A V T ++ + +++ + G+ L +G F A
Sbjct 215 QKGAPTTLNVGGMAPVTTKFRDVPNLSGTPLIFRDNKGRTIKTGQ--LGIGPVDAGFLVA 272
Query 177 AFSGTVAAG--LYPNNLYADLSSANAISVDDLRLAFAYQKMLERDAIYGSRYNEYLYGHF 234
+ A G P+NL+ADLS+A IS+ DLRLA YQ E DA G+RY E+ HF
Sbjct 273 QNTAQAANGERAIPSNLWADLSNATGISISDLRLAITYQHYKEMDARGGTRYVEFTLNHF 332
Query 235 GVHIPDAYIQFPQYLGGGRTPLNIVQVAQTSQGTEE-SPLGNVGAYSWTNGRTGY--SRK 291
GVH DA +Q ++LGG L + V QTS E+ +P GN+ A+S T + Y ++
Sbjct 333 GVHTADARLQRSEFLGGHSQSLLVQSVPQTSSTVEKMTPQGNLAAFSETMIQNNYLVNKT 392
Query 292 FKEHGIVMTVACLRYRHTYQQGIAKKW-RRKVREDFYDPLFSTIGQQPVYTTELYAQAFP 350
F EH ++ +A +RY+HTYQQGI W R + + D YDPL + I +QPV E+ Q
Sbjct 393 FTEHSYIIVLAVVRYKHTYQQGIEADWFRGQDKFDMYDPLLANISEQPVKNREIMVQGNS 452
Query 351 Q--TVFGYREAWSELRNIPNTISGEMRSGVTNSLDIWHFADNYSSSPTLSQSFTEETPSY 408
Q +FG++EAW++LR PN+++G MRS SLD WHFAD+Y+ P LS + +E
Sbjct 453 QDNEIFGFQEAWADLRFKPNSVAGVMRSSHPQSLDYWHFADHYAQLPKLSSEWLKEDYKN 512
Query 409 VDRTLSVPSSSQN-NFILNFYFDMSAVRKMPVYSMPSL 445
VDRTL++ +S ++F F+ A + MP+YS P L
Sbjct 513 VDRTLALKASDNTPQLRVDFMFNTIAEKPMPLYSTPGL 550
>gi|530695351|gb|AGT39907.1| major capsid protein [Marine gokushovirus]
Length=539
Score = 269 bits (687), Expect = 5e-80, Method: Compositional matrix adjust.
Identities = 174/466 (37%), Positives = 258/466 (55%), Gaps = 50/466 (11%)
Query 4 NPNPSAYVNNVLEEIPMTYGEVNPGSIADYLGLPI-GSISQSSPVS--VLPFRCFALIYD 60
+P+P + ++ + + G S+ DY+GLP G + S +S L R + LI++
Sbjct 102 DPDPDSSIDYTIPTMTSPNGGYAVNSLQDYMGLPTAGQVDAGSSISHNSLFTRAYNLIWN 161
Query 61 KYFRNENTTDEIYIQKKGFSLSELIGAQNFSPNSYCG-KLPKVNKYKDYFTSCVPNPQKG 119
++FR+EN D + + K P++Y L + K DYFTS +P PQKG
Sbjct 162 EWFRDENLQDSVVVDKGD------------GPDTYTDYTLLRRGKRHDYFTSALPWPQKG 209
Query 120 APVTFNLGDQAVVRTSDS-------ELVTGPQEQMALTNSQSGSASVGEHPLIVGLGGMR 172
VT LG A V +D+ E+ TG S S A+ G M
Sbjct 210 DAVTLPLGGSANVVYNDTGDPAYIREVSTGNVWTTPSRESVSKEAN----------GNM- 258
Query 173 FDAAAFSGTVAAGLYPN-NLYADLSSANAISVDDLRLAFAYQKMLERDAIYGSRYNEYLY 231
+ +G+V A PN +L ADLS+A A +++ +R +F Q++LERDA G+RY E +
Sbjct 259 ---SVPTGSVNAQYDPNGSLVADLSTATAATINAIRQSFQIQRLLERDARGGTRYTEIVR 315
Query 232 GHFGVHIPDAYIQFPQYLGGGRTPLNIVQVAQTS----QGTEESPLGNVGAY--SWTNGR 285
HFGV PDA +Q P+YLGGG P+ + VAQ S GT ++PLG +GA +G
Sbjct 316 SHFGVISPDARMQRPEYLGGGSAPIIVNPVAQQSASGASGT-DTPLGTLGAVGTGLASGH 374
Query 286 TGYSRKFKEHGIVMTVACLRYRHTYQQGIAKKWRRKVREDFYDPLFSTIGQQPVYTTELY 345
G++ F EHG+V+ + +R TYQQG+ + + R R DF+ P+FS +G+QP+ ELY
Sbjct 375 -GFASSFTEHGVVVGLCSVRADLTYQQGLHRMFSRSTRYDFFFPVFSHLGEQPILNKELY 433
Query 346 A--QAFPQTVFGYREAWSELRNIPNTISGEMRSGVTNSLDIWHFADNYSSSPTLSQSFTE 403
A + VFGY+EAW+E R P+ ++G MRS +LD WH A N+ S PTL+ +F E
Sbjct 434 ATGTSTDDDVFGYQEAWAEYRYKPSQVTGLMRSTAAGTLDAWHLAQNFGSLPTLNSTFIE 493
Query 404 ETPSYVDRTLSVPSSSQ-NNFILNFYFDMSAVRKMPVYSMPSLIDH 448
+TP VDR ++V S + FI + +FD++ R MP+YS+P L+DH
Sbjct 494 DTPP-VDRVVAVGSEANGQQFIFDAFFDINMARPMPMYSVPGLVDH 538
Lambda K H a alpha
0.317 0.134 0.400 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3028457194728