bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-24_CDS_annotation_glimmer3.pl_2_4
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
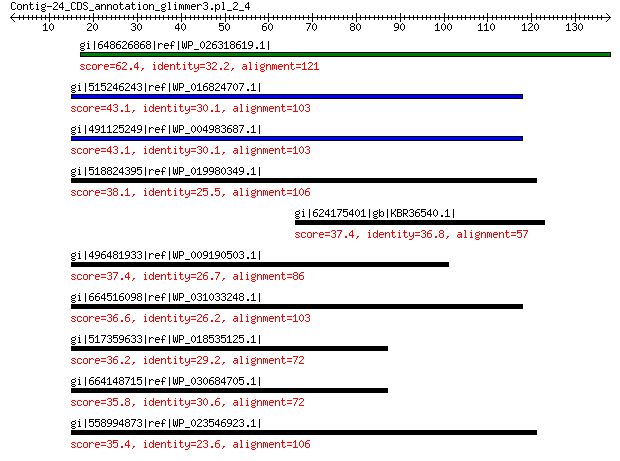
gi|648626868|ref|WP_026318619.1| hypothetical protein 62.4 5e-10
gi|515246243|ref|WP_016824707.1| peptidase 43.1 0.019
gi|491125249|ref|WP_004983687.1| peptidase 43.1 0.019
gi|518824395|ref|WP_019980349.1| peptidase 38.1 0.98
gi|624175401|gb|KBR36540.1| hypothetical protein X418_00653 37.4 1.5
gi|496481933|ref|WP_009190503.1| peptidase 37.4 1.8
gi|664516098|ref|WP_031033248.1| peptidase 36.6 3.9
gi|517359633|ref|WP_018535125.1| peptidase 36.2 5.4
gi|664148715|ref|WP_030684705.1| hypothetical protein 35.8 5.9
gi|558994873|ref|WP_023546923.1| peptidase 35.4 8.2
>gi|648626868|ref|WP_026318619.1| hypothetical protein [Alistipes onderdonkii]
Length=101
Score = 62.4 bits (150), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 59/121 (49%), Gaps = 21/121 (17%)
Query 17 NLSQRVGIRSCADQHASYRTRNCASRLDEFIVGARGMNEILEEYYTFGFLSCDTQAVRGD 76
+L + V + SC++ D I G + + + L+EY+ G CD
Sbjct 2 DLVESVVVHSCSE--------------DCLIHGVKSLRQGLQEYWETGVYPCDPVQRNSG 47
Query 77 SAYDEIQPSGKDASFLSTDPSSDFSLDKFERIERIAECVGETSAERHKEELGKQNDKQND 136
YDE S DPS+DFSLDKFER+E+I E V E ++H+++LG D++
Sbjct 48 EYYDE-------DSTTQVDPSTDFSLDKFERLEKIVEVVSERERKKHEDDLGVTPDEELK 100
Query 137 K 137
K
Sbjct 101 K 101
>gi|515246243|ref|WP_016824707.1| peptidase [Streptomyces viridosporus]
Length=523
Score = 43.1 bits (100), Expect = 0.019, Method: Composition-based stats.
Identities = 31/106 (29%), Positives = 48/106 (45%), Gaps = 12/106 (11%)
Query 15 YSNLSQRVGIRSCADQHASYRTRNCASRLDEFIVGARGMNEILEEYYTFGFLSCDTQAVR 74
YSNL +CAD YRT + L EF R +E+ EY +G + C AV
Sbjct 376 YSNLVPANVSINCADDKPRYRTEDVERSLPEF----RAASELFGEYLAWGLIGCTDWAVP 431
Query 75 GDSAYDEIQPSGKDASFL---STDPSSDFSLDKFERIERIAECVGE 117
G + + E+ G + + DP++ +E R+A+ +GE
Sbjct 432 GAADHPEVSAPGAAPILVIGNTGDPAT-----PYEGARRMAQALGE 472
>gi|491125249|ref|WP_004983687.1| peptidase [Streptomyces ghanaensis]
gi|291340295|gb|EFE67251.1| peptidase [Streptomyces ghanaensis ATCC 14672]
Length=523
Score = 43.1 bits (100), Expect = 0.019, Method: Composition-based stats.
Identities = 31/106 (29%), Positives = 48/106 (45%), Gaps = 12/106 (11%)
Query 15 YSNLSQRVGIRSCADQHASYRTRNCASRLDEFIVGARGMNEILEEYYTFGFLSCDTQAVR 74
YSNL +CAD YRT + L EF R +E+ EY +G + C AV
Sbjct 376 YSNLVPANVSINCADDKPRYRTEDVERSLPEF----RAASELFGEYLAWGLIGCTDWAVP 431
Query 75 GDSAYDEIQPSGKDASFL---STDPSSDFSLDKFERIERIAECVGE 117
G + + E+ G + + DP++ +E R+A+ +GE
Sbjct 432 GAADHPEVSAPGAAPILVIGNTGDPAT-----PYEGARRMAQALGE 472
>gi|518824395|ref|WP_019980349.1| peptidase [Streptomyces sp. Amel2xE9]
Length=519
Score = 38.1 bits (87), Expect = 0.98, Method: Composition-based stats.
Identities = 27/109 (25%), Positives = 51/109 (47%), Gaps = 12/109 (11%)
Query 15 YSNLSQRVGIRSCADQHASYRTRNCASRLDEFIVGARGMNEILEEYYTFGFLSCDTQAVR 74
YSN++ +CAD+ Y T +L EF R + + E+ +G LSC V+
Sbjct 379 YSNITAANVAINCADEKQRYDTAYVRRKLPEF----RSASPLFGEFLAWGMLSCANWPVK 434
Query 75 GDSAYDEIQPSGKDASFL---STDPSSDFSLDKFERIERIAECVGETSA 120
G + + ++ SG + + DP++ +E ++A+ +G+ A
Sbjct 435 GAADHPDVSASGSAPVLVVGNTGDPAT-----PYEGARKMADALGKGVA 478
>gi|624175401|gb|KBR36540.1| hypothetical protein X418_00653 [Mycobacterium tuberculosis XTB13-212]
Length=373
Score = 37.4 bits (85), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 21/57 (37%), Positives = 32/57 (56%), Gaps = 2/57 (4%)
Query 66 LSCDTQAVRGDSAYDEIQPSGKDASFLSTDPSSDFSLDKFERIERIAECVGETSAER 122
L D ++RG A E P+ +D +F++ DP+S F+LD+ E + E V SA R
Sbjct 32 LGADAASIRG--AVSEQSPNYRDGAFVNLDPASMFTLDREELRLIVWELVARHSASR 86
>gi|496481933|ref|WP_009190503.1| peptidase [Streptomyces sp. e14]
gi|292834607|gb|EFF92956.1| proteinase [Streptomyces sp. e14]
Length=513
Score = 37.4 bits (85), Expect = 1.8, Method: Composition-based stats.
Identities = 23/89 (26%), Positives = 42/89 (47%), Gaps = 7/89 (8%)
Query 15 YSNLSQRVGIRSCADQHASYRTRNCASRLDEFIVGARGMNEILEEYYTFGFLSCDTQAVR 74
YSN++ +CAD+ Y T +L EF R + + E+ +G LSC V+
Sbjct 373 YSNITAANVAINCADEKQRYDTAYVQRKLPEF----RSASPLFGEFLAWGMLSCANWPVK 428
Query 75 GDSAYDEIQPSGKDASFL---STDPSSDF 100
G + + ++ SG + + DP++ +
Sbjct 429 GAADHPDVSASGSAPVLVVGNTGDPATPY 457
>gi|664516098|ref|WP_031033248.1| peptidase [Streptomyces olivaceus]
Length=519
Score = 36.6 bits (83), Expect = 3.9, Method: Composition-based stats.
Identities = 27/106 (25%), Positives = 48/106 (45%), Gaps = 12/106 (11%)
Query 15 YSNLSQRVGIRSCADQHASYRTRNCASRLDEFIVGARGMNEILEEYYTFGFLSCDTQAVR 74
YSN+ SCAD A Y + + RL EF R + + +Y + +SC AV
Sbjct 379 YSNIGAANTAISCADDKARYDSADVEGRLPEF----RKASPVFGDYLAWSMVSCTDWAVP 434
Query 75 GDSAYDEIQPSGKDASFL---STDPSSDFSLDKFERIERIAECVGE 117
G + + ++ G + + DP++ +E ++AE +G+
Sbjct 435 GAAGHPDVGAPGAPPILVIGNTGDPAT-----PYEGAGKMAEALGK 475
>gi|517359633|ref|WP_018535125.1| peptidase [Streptomyces sp. HmicA12]
Length=515
Score = 36.2 bits (82), Expect = 5.4, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 35/72 (49%), Gaps = 4/72 (6%)
Query 15 YSNLSQRVGIRSCADQHASYRTRNCASRLDEFIVGARGMNEILEEYYTFGFLSCDTQAVR 74
YSN+ +CAD Y ++ ++L EF R + + +Y +G LSC AV
Sbjct 375 YSNMQAANVAINCADYKPRYTAKDVEAKLPEF----RKASPLFGDYLAWGMLSCTGWAVD 430
Query 75 GDSAYDEIQPSG 86
G + + ++ SG
Sbjct 431 GAADHPDVSASG 442
>gi|664148715|ref|WP_030684705.1| hypothetical protein, partial [Streptomyces sp. NRRL B-1347]
Length=517
Score = 35.8 bits (81), Expect = 5.9, Method: Composition-based stats.
Identities = 22/72 (31%), Positives = 33/72 (46%), Gaps = 4/72 (6%)
Query 15 YSNLSQRVGIRSCADQHASYRTRNCASRLDEFIVGARGMNEILEEYYTFGFLSCDTQAVR 74
YSNLS +CAD Y T + +L F R + + +Y +G L C AV
Sbjct 378 YSNLSAANVAINCADDKPRYTTADVEEKLPRF----REASPLFGDYLAWGMLGCTDWAVP 433
Query 75 GDSAYDEIQPSG 86
G + + ++ SG
Sbjct 434 GQADHPDVSASG 445
>gi|558994873|ref|WP_023546923.1| peptidase [Streptomyces roseochromogenus]
gi|558541347|gb|EST32426.1| peptidase [Streptomyces roseochromogenes subsp. oscitans DS 12.976]
Length=514
Score = 35.4 bits (80), Expect = 8.2, Method: Composition-based stats.
Identities = 25/109 (23%), Positives = 50/109 (46%), Gaps = 12/109 (11%)
Query 15 YSNLSQRVGIRSCADQHASYRTRNCASRLDEFIVGARGMNEILEEYYTFGFLSCDTQAVR 74
YSN++ +CAD+ Y + +L EF R + + +Y +G +SC AV
Sbjct 374 YSNITAANTAINCADEKPRYTADDVQRKLPEF----RAASPLFGDYLAWGMVSCTDWAVP 429
Query 75 GDSAYDEIQPSGKDASFL---STDPSSDFSLDKFERIERIAECVGETSA 120
G + + ++ G + + DP++ +E +++ E +G+ A
Sbjct 430 GAADHPDVSAPGSAPILVVGNTGDPAT-----PYEGAKKMVEALGKGVA 473
Lambda K H a alpha
0.315 0.131 0.381 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 439745511555