bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_8
Length=262
Score E
Sequences producing significant alignments: (Bits) Value
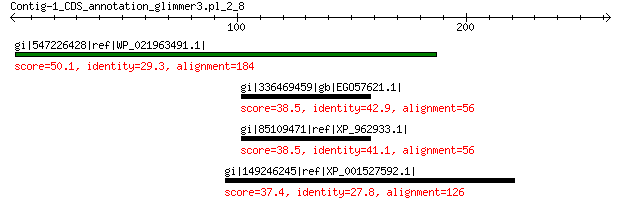
gi|547226428|ref|WP_021963491.1| putative uncharacterized protein 50.1 8e-04
gi|336469459|gb|EGO57621.1| hypothetical protein NEUTE1DRAFT_122010 38.5 4.9
gi|85109471|ref|XP_962933.1| hypothetical protein NCU06217 38.5 5.1
gi|149246245|ref|XP_001527592.1| conserved hypothetical protein 37.4 9.3
>gi|547226428|ref|WP_021963491.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103380|emb|CCY83991.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=416
Score = 50.1 bits (118), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 54/213 (25%), Positives = 92/213 (43%), Gaps = 47/213 (22%)
Query 3 RQDFSGLSNTLASALQISNQTKETNANVQTLQSQKSLYDAQANSILSNVDWWKLGPEYK- 61
R DFSG++ + + L I Q+QK + DAQA S+ +K+ +YK
Sbjct 152 RPDFSGVTGIIQTLLDI--------------QAQKGVRDAQAFSLGEQASGFKIENKYKA 197
Query 62 ----------------KWSQMT----GLARAGLQFQTD----KQNLRNMTWSGNLIQAQ- 96
K SQ + AR F +D ++ N ++G LI+AQ
Sbjct 198 EKLLWDIYNSKADYNLKNSQESLNNMSFARLQAMFSSDVSKAQREAENAQFTGELIRAQT 257
Query 97 ---HIGALLDNKSKRIINNYLDEGQRLQLDLMAAQYYDAMASGHLKYQQAKSEITKRILM 153
+ LL K + Y D+ +L +M+AQ Y +A+G QA+ I + +
Sbjct 258 ACQQLQGLLGAKELK----YYDQKVLQELAIMSAQQYSLVAAGKASEAQARQAIENALNL 313
Query 154 MAEAKGLQINNKAAEDTADGYIKALNAEYSASY 186
+ + +G++++N + TA+ IK + SY
Sbjct 314 VEQREGIKVDNYVKQKTANALIKTARNNCNTSY 346
>gi|336469459|gb|EGO57621.1| hypothetical protein NEUTE1DRAFT_122010 [Neurospora tetrasperma
FGSC 2508]
gi|350290896|gb|EGZ72110.1| RNA-binding domain-containing protein [Neurospora tetrasperma
FGSC 2509]
Length=771
Score = 38.5 bits (88), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 24/56 (43%), Positives = 36/56 (64%), Gaps = 3/56 (5%)
Query 102 LDNKSKRIINNYLDEGQRLQLDLMAAQYYDAMASGHLKYQQAKSEITKRILMMAEA 157
L+ K+K +NNYL EG+RL+LD+ A++ DA +G + + A E KR +AEA
Sbjct 100 LEAKAK--LNNYLLEGKRLKLDIAEARHRDAKKTGPVVSKVA-EEKQKRAEAVAEA 152
>gi|85109471|ref|XP_962933.1| hypothetical protein NCU06217 [Neurospora crassa OR74A]
gi|28924577|gb|EAA33697.1| nucleolar protein 4 [Neurospora crassa OR74A]
Length=772
Score = 38.5 bits (88), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 35/56 (63%), Gaps = 3/56 (5%)
Query 102 LDNKSKRIINNYLDEGQRLQLDLMAAQYYDAMASGHLKYQQAKSEITKRILMMAEA 157
L+ K+K +NNYL EG+RL+LD+ A++ DA +G + + E KR +AEA
Sbjct 100 LEAKAK--LNNYLLEGKRLKLDIAEARHRDAKKTGPV-VSKVAEEKQKRAEAVAEA 152
>gi|149246245|ref|XP_001527592.1| conserved hypothetical protein [Lodderomyces elongisporus NRRL
YB-4239]
gi|146447546|gb|EDK41934.1| conserved hypothetical protein [Lodderomyces elongisporus NRRL
YB-4239]
Length=660
Score = 37.4 bits (85), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 35/128 (27%), Positives = 64/128 (50%), Gaps = 8/128 (6%)
Query 95 AQHIGALLDNKSK-RIINNYLDEGQRLQLDLMAAQYYDAMASGHLKYQQAKSEITKRILM 153
AQ I A LD +K R++ N ++E +L+ ++A++ + LK + +++++R+L
Sbjct 474 AQRIEAQLDQVAKSRVVLNSINE----KLNTLSAKHDLGNTTRILKAKARHTQLSRRVLR 529
Query 154 MAEAKG-LQINNKAAEDTADGYIKALNAEYSASYDINSPFKYGEDSYVPASVLKSRMDAL 212
+A L++ +G K + S D +SP D + +VLK R + L
Sbjct 530 LATVLAILKLKGYPLLPEEEGIAKQFDLLSSKINDPSSPIGKLGDIFAKLTVLKERAEDL 589
Query 213 NSKWQFDK 220
NS QFD+
Sbjct 590 NS--QFDQ 595
Lambda K H a alpha
0.313 0.127 0.364 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1223431011738