bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-15_CDS_annotation_glimmer3.pl_2_5
Length=489
Score E
Sequences producing significant alignments: (Bits) Value
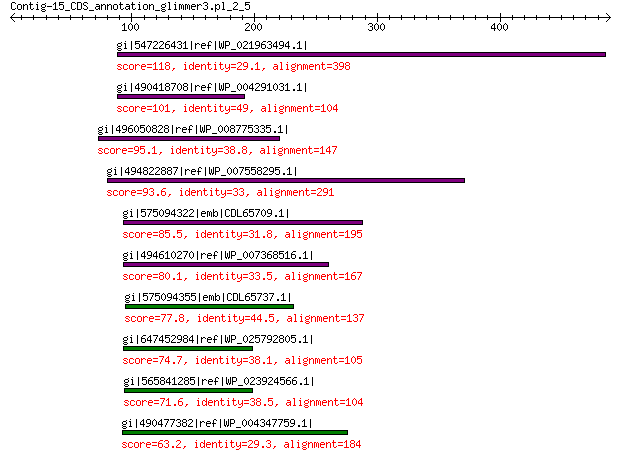
gi|547226431|ref|WP_021963494.1| predicted protein 118 1e-25
gi|490418708|ref|WP_004291031.1| hypothetical protein 101 6e-20
gi|496050828|ref|WP_008775335.1| hypothetical protein 95.1 1e-17
gi|494822887|ref|WP_007558295.1| hypothetical protein 93.6 4e-17
gi|575094322|emb|CDL65709.1| unnamed protein product 85.5 2e-14
gi|494610270|ref|WP_007368516.1| hypothetical protein 80.1 7e-13
gi|575094355|emb|CDL65737.1| unnamed protein product 77.8 4e-12
gi|647452984|ref|WP_025792805.1| hypothetical protein 74.7 4e-11
gi|565841285|ref|WP_023924566.1| hypothetical protein 71.6 4e-10
gi|490477382|ref|WP_004347759.1| hypothetical protein 63.2 3e-07
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 118 bits (296), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 116/404 (29%), Positives = 181/404 (45%), Gaps = 41/404 (10%)
Query 88 HLQDYYGYISRKDGQLFVKRLRKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDS 147
+L Y Y S++D QLF+KR+RK++ K++ EK+ Y+VSEYGP FR H+H+L F D
Sbjct 125 NLDGYLSYTSKRDAQLFLKRVRKNLSKYSD--EKIRYYIVSEYGPKTFRAHYHVLFFYDE 182
Query 148 DRVAKNIGRIVNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFS 207
+ K + +++ +W+FGR D S SRG SYVA YVN LP L D KP+S S
Sbjct 183 VKTQKVMSKVIRQAWQFGRVDCSLSRGKCNSYVARYVNCNYCLPRFLG-DMSTKPFSCHS 241
Query 208 NGFATSCFFDAKEAV-RDSISRPIEETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLR 266
FA KE + + S+ I ++ ING + P R++ SC
Sbjct 242 IRFALGIHQSQKEEIYKGSVDDFIYQS----------GEINGNYVEFMPWRNL--SCTFF 289
Query 267 YACNG--RLSSHEL---YWLVRSVSTTLSRSFQTVRRDNPDATLMDVCRLHLRSIYSMSS 321
C G R S EL Y ++R V + + SF T+ + ++D L + + +S S
Sbjct 290 PKCKGYSRKSDSELWQSYNILREVRSAIGYSFNTII--DYARCILD---LVVTAKFSCDS 344
Query 322 RSVEDFLSIENQLYTCYYYARLESPTTDVRSDDLFDSDCMRLYRLFSCVGKFISFWDITP 381
R + N++ + Y T SD L D + R F++F +
Sbjct 345 RGLPCSSPALNKVIS---YFSQGIDTNPYYSDYLADYHTNSIARELYISRHFLTF--VCD 399
Query 382 SSSYEYIYRTVDLSKWYYSRLSYHMLRSQYTdlvglvdldeelvdFLIAPVTEDTPSSNG 441
+ SY YR L + ++ R Y +L YT + L + I D+ +
Sbjct 400 NDSYHERYRKFTLIRQFWQRYDYALLVGMYTSQIENRHLISNYDWYYINKTPLDSFGNVD 459
Query 442 VSALEHHPPFESLVKSHPILSLAQAKAKTDCDNRVKHREINERN 485
VS L ++ V K+ + + +KH+ N+ N
Sbjct 460 VSQLSKELFYKRFV----------IKSDENFEKSIKHKIQNDAN 493
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 101 bits (251), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 51/104 (49%), Positives = 65/104 (63%), Gaps = 1/104 (1%)
Query 88 HLQDYYGYISRKDGQLFVKRLRKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDS 147
HL Y Y+ + D QLF KR R ++ K K EKV + + EYGPVHFRPH+HILLF S
Sbjct 36 HLFGYLPYLRKFDLQLFFKRFRYYVAKRFPK-EKVRYFAIGEYGPVHFRPHYHILLFLQS 94
Query 148 DRVAKNIGRIVNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLP 191
D + ++V+ +W FGR D S+G SYVAGYVNS +P
Sbjct 95 DEALQVCSKVVSEAWPFGRVDCQLSKGKCSSYVAGYVNSSVLVP 138
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 95.1 bits (235), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 57/147 (39%), Positives = 76/147 (52%), Gaps = 4/147 (3%)
Query 73 DFREQATLNVNGKYPHLQDYYGYISRKDGQLFVKRLRKHIFKFAGKYEKVHIYLVSEYGP 132
D E N+ K+ D Y+ + D QLF+KRLR ++ K EKV + V EYGP
Sbjct 98 DLTEDERTNLLNKFYLFGDV-PYLRKTDLQLFLKRLRYYVTK-QKPSEKVRYFAVGEYGP 155
Query 133 VHFRPHFHILLFTDSDRVAKNIGRIVNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLPY 192
VHFRPH+H+LLF SD + ++ +W FGR D S+G +YVA YVNS +P
Sbjct 156 VHFRPHYHLLLFLQSDEALQICSENISKAWTFGRVDCQVSKGQCSNYVASYVNSSCTIPK 215
Query 193 HLKQDDRVKPYSRFSNGFATSCFFDAK 219
K V P+S S F D +
Sbjct 216 VFKASS-VCPFSVHSQKLGQG-FLDCQ 240
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 93.6 bits (231), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 96/331 (29%), Positives = 142/331 (43%), Gaps = 74/331 (22%)
Query 80 LNVNGKYPHLQDYYGYISRKDGQLFVKRLRKHIFKFAGKYEKVHIYLVSEYGPVHFRPHF 139
+N G++P Y+S+++ QLF+KRLRK++ K+ G +K+ + EYGP+ FRPHF
Sbjct 112 INYKGRFP-------YLSKRELQLFMKRLRKYLDKYEG--QKIRFFATGEYGPLSFRPHF 162
Query 140 HILLFTD-----------------------------SDRVAKNIGRIVNSSWKFGRCD-W 169
HILLF D + + + SW FG D
Sbjct 163 HILLFVDDPSLFLPSVHTLGEYPYPYWSKYQKAHCGKGTLLSKLEYYIRESWPFGGIDAQ 222
Query 170 SASRGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSNGFATSCFFDAKEAVRDSISRP 229
S +G SYVAGYVNS LP LK D VK +S+ S F E + P
Sbjct 223 SVEQGSCSSYVAGYVNSSVPLPSCLKVDA-VKSFSQHSRFLGRKIF--GTELI------P 273
Query 230 IEETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNG-RLSSHELYWLVRSVSTT 288
+ + + F+ G+ R P ++ S + + C G L SHE + V ++ +
Sbjct 274 LLKLKFTEFVQR-SFFCRGRYDNFRTPSEMLHSVYPQ--CKGFALLSHEQRFRVYTIWSR 330
Query 289 LSRSFQTVRRDNPDATLMDVCRLHLRSIYS--------MSSRSVEDFLSIENQLYTCYYY 340
L F + ++ DV R + S YS + R EDFL I +L Y
Sbjct 331 LRYYFNSDKK-------ADVARSLVTSFYSWLDTGILRVPERVREDFLLIYTELSQNLNY 383
Query 341 ARLESPTTD-VRSDDLFDSDCMRLYRLFSCV 370
R++ D R DD ++ +LF CV
Sbjct 384 KRIDRFDYDKFRHDDDLNN------QLFQCV 408
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 85.5 bits (210), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 62/210 (30%), Positives = 94/210 (45%), Gaps = 26/210 (12%)
Query 93 YGYISRKDGQLFVKRLRKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAK 152
Y + +D QLF+KRLRKHI+K+ G EK+ Y++ EYG RPH+H LLF +S +++
Sbjct 139 YALLYYRDIQLFLKRLRKHIYKYYG--EKIRFYIIGEYGTKSLRPHWHCLLFFNSSSLSQ 196
Query 153 ------NIG---------RIVNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLPYHLKQD 197
N+G R + W+FG CD + G+A +YV+ YVN + P L
Sbjct 197 AFEDCVNVGTTSRPCSCPRFLRPFWQFGICDSKRTNGEAYNYVSSYVNQSANFPKLLVLL 256
Query 198 DRVKPYSRFSNGFATSCFFDAKEAVRDSISRPIEETPLSPFLNGVPSLINGKLLAIRPPR 257
K Y G S SI I++ S F G + R
Sbjct 257 SNQKAYHSIQLGQILS---------EQSIVSAIQKGDFSFFERQFYLDTFGAANSYSVWR 307
Query 258 SVVDSCFLRYACNGRLSSHELYWLVRSVST 287
S F ++ C+ +L+ + Y ++ T
Sbjct 308 SYYSRFFPKFTCSSQLTYEQTYRVLTCYET 337
>gi|494610270|ref|WP_007368516.1| hypothetical protein [Prevotella multiformis]
gi|324988542|gb|EGC20505.1| hypothetical protein HMPREF9141_0984 [Prevotella multiformis
DSM 16608]
Length=479
Score = 80.1 bits (196), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 56/176 (32%), Positives = 84/176 (48%), Gaps = 22/176 (13%)
Query 93 YGYISRKDGQLFVKRLRK----HIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSD 148
+ Y +KD Q + KRLR + K ++ ++ SEYGP FRPH+H +L+ DS+
Sbjct 116 FAYPCKKDVQDWFKRLRSAVDYQLNKNKSNEFRIRYFICSEYGPRTFRPHYHAILWYDSE 175
Query 149 RVAKNIGRIVNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSN 208
+ +NIGR++ +WK G +S A YVA YVN +RLP L+ +
Sbjct 176 ELQRNIGRLIRETWKNGNSVFSLVNNSASQYVAKYVNGDTRLPPFLRTE----------- 224
Query 209 GFATSCFFDAKEAVRDSISRPIEETPLSPFLNG-----VPSLINGKLLAIRPPRSV 259
TS F A + + EE S L+G V + NG+ + PRS+
Sbjct 225 --FTSTFHLASKHPYIGYCKADEEALRSNVLDGTYGQSVLNRDNGQFEFVPTPRSL 278
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 77.8 bits (190), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 61/170 (36%), Positives = 83/170 (49%), Gaps = 36/170 (21%)
Query 95 YISRKDGQLFVKRLRKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNI 154
Y+ ++D QLF+KRLRK++ K++ KV + + EYGPVHFRPH+H LLF D +
Sbjct 125 YLRKRDLQLFIKRLRKNLSKYSDA--KVRYFAMGEYGPVHFRPHYHFLLFFDEIKFTAPS 182
Query 155 GRI----------------------------VNSSWKFGRCDWSASRGDAESYVAGYVNS 186
G + SSWKFGR D S+GDA YV+ YV+
Sbjct 183 GHTLGEFPDWAWYDSQNKCSRSDILSVVEYCIRSSWKFGRVDAQYSKGDAAQYVSSYVSG 242
Query 187 FSRLPYHLKQDDRVKPY---SRF-SNGF-ATSCFFDAKEAVRDSISRPIE 231
LP + Q +P+ SRF GF A C + VRD + R +E
Sbjct 243 SGSLP-KVYQVSSARPFSLHSRFLGQGFLAHECEKVYETPVRDFVKRSVE 291
>gi|647452984|ref|WP_025792805.1| hypothetical protein [Prevotella histicola]
Length=480
Score = 74.7 bits (182), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 40/108 (37%), Positives = 58/108 (54%), Gaps = 3/108 (3%)
Query 93 YGYISRKDGQLFVKRLRKHI---FKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDR 149
+ Y +KD Q F KRLR I K G ++ ++ SEYGP FRPH+H +L+ DS+
Sbjct 119 FAYPCKKDVQDFFKRLRSKIDYKLKPRGNEYRIRYFICSEYGPNTFRPHYHAILWYDSEI 178
Query 150 VAKNIGRIVNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLPYHLKQD 197
+ + ++ +WK G D+S A YVA YVN LP L+ +
Sbjct 179 LHNELNVLIRETWKNGNTDFSLVNSSASQYVAKYVNGDCDLPSFLRTE 226
>gi|565841285|ref|WP_023924566.1| hypothetical protein [Prevotella nigrescens]
gi|564729906|gb|ETD29850.1| hypothetical protein HMPREF1173_00032 [Prevotella nigrescens
CC14M]
Length=484
Score = 71.6 bits (174), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 40/109 (37%), Positives = 60/109 (55%), Gaps = 5/109 (5%)
Query 94 GYISRKDGQLFVKRLRKHIFKFAGKY-----EKVHIYLVSEYGPVHFRPHFHILLFTDSD 148
Y + D F KRLR + + K+ EK+ ++ SEYGP RPH+H +++ DS+
Sbjct 113 AYCCKSDIVKFFKRLRSKLSYYFKKHHIITNEKIRYFVCSEYGPKTLRPHYHAIIWFDSE 172
Query 149 RVAKNIGRIVNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLPYHLKQD 197
VA+ I ++++SSW G D+ A YVA YV+ S LP L+ D
Sbjct 173 EVARVIEKMLSSSWSNGFTDFEYVNSTAPQYVAKYVSGNSVLPEILQHD 221
>gi|490477382|ref|WP_004347759.1| hypothetical protein [Prevotella buccalis]
gi|281300711|gb|EFA93042.1| hypothetical protein HMPREF0650_1078 [Prevotella buccalis ATCC
35310]
Length=582
Score = 63.2 bits (152), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 54/208 (26%), Positives = 88/208 (42%), Gaps = 28/208 (13%)
Query 92 YYGYISRKDGQLFVKRLRKHIFKFAGKYE---KVHIYLVSEYGPVHFRPHFHILLFTDSD 148
++G + +KD Q F+KRLR I K + K+ Y+ SEYGP +RPH+H +LF DS
Sbjct 137 HFGVVCKKDIQNFLKRLRWRISKIPNITKDESKIRYYISSEYGPTTYRPHYHGILFFDSK 196
Query 149 RVAKNIGRIVNSSW-KFGRCDWSASR--------------------GDAESYVAGYVNSF 187
++ I ++ SW K+ R +R + YVA YV
Sbjct 197 KILDKIKSLIVMSWGKYERQQGERNRFKFRPFARPSLTSDYIKLCDPNTAYYVASYVAGN 256
Query 188 SRLPYHLKQDDRVKPYSRFSNGFATSCFFDAKEAVRDSISRPIEETPLSPFLNGVPSLIN 247
LP L Q KP+ S + K+ + ++I+R T F + +
Sbjct 257 DNLPQVL-QLRETKPFHVQSKNPVIGSYKVDKQEILENINRGTYTTTKQVFDDKTGQFND 315
Query 248 GKLLAIRPPRSVVDSCFLRYACNGRLSS 275
+ + P + + S F + C L++
Sbjct 316 ---VIVPLPETTLSSIFRKCVCYSTLTN 340
Lambda K H a alpha
0.322 0.137 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3408282129351