bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
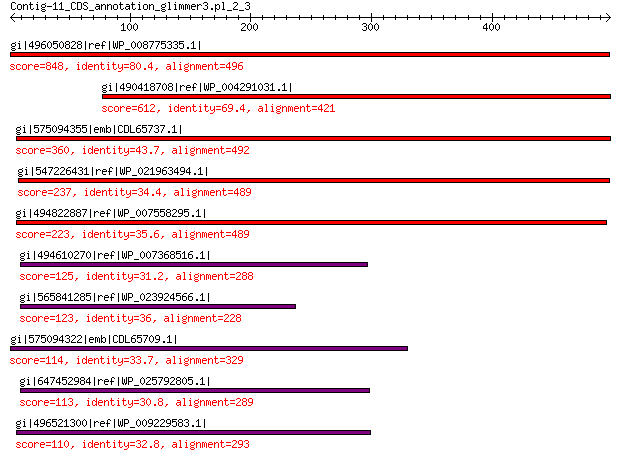
Query= Contig-11_CDS_annotation_glimmer3.pl_2_3
Length=497
Score E
Sequences producing significant alignments: (Bits) Value
gi|496050828|ref|WP_008775335.1| hypothetical protein 848 0.0
gi|490418708|ref|WP_004291031.1| hypothetical protein 612 0.0
gi|575094355|emb|CDL65737.1| unnamed protein product 360 6e-115
gi|547226431|ref|WP_021963494.1| predicted protein 237 8e-68
gi|494822887|ref|WP_007558295.1| hypothetical protein 223 3e-62
gi|494610270|ref|WP_007368516.1| hypothetical protein 125 6e-28
gi|565841285|ref|WP_023924566.1| hypothetical protein 123 3e-27
gi|575094322|emb|CDL65709.1| unnamed protein product 114 4e-24
gi|647452984|ref|WP_025792805.1| hypothetical protein 113 1e-23
gi|496521300|ref|WP_009229583.1| hypothetical protein 110 2e-22
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 848 bits (2192), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 399/497 (80%), Positives = 448/497 (90%), Gaps = 1/497 (0%)
Query 1 MIKNSFCKCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFIT 60
M++N FCKCLHPK+I+NPYT ESMVVPCGHCQAC LAKNSRYAFQCDLESY AKHTLFIT
Sbjct 1 MVQNPFCKCLHPKRIMNPYTKESMVVPCGHCQACTLAKNSRYAFQCDLESYTAKHTLFIT 60
Query 61 LTYANRYIPRATFVDSLERPFGNDLVDKETGEILGPSDMKQEDIDRLLNKFYLFGDVPYL 120
LTYANR+IPRA FVDS+ERP+G DL+DKETGEILGP+D+ +++ LLNKFYLFGDVPYL
Sbjct 61 LTYANRFIPRAMFVDSIERPYGCDLIDKETGEILGPADLTEDERTNLLNKFYLFGDVPYL 120
Query 121 RKTDLQLFFKRLRYYVSKQCPSEKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQVCSEN 180
RKTDLQLF KRLRYYV+KQ PSEKVRYFAVGEYGPVHFRPHYH+LLFLQSDEALQ+CSEN
Sbjct 121 RKTDLQLFLKRLRYYVTKQKPSEKVRYFAVGEYGPVHFRPHYHLLLFLQSDEALQICSEN 180
Query 181 ISQAWTLGRIDCQISKGQCSSYVASYVNSSCTIPKVFKLSSVCPFNVHSQKLGQGFLDCQ 240
IS+AWT GR+DCQ+SKGQCS+YVASYVNSSCTIPKVFK SSVCPF+VHSQKLGQGFLDCQ
Sbjct 181 ISKAWTFGRVDCQVSKGQCSNYVASYVNSSCTIPKVFKASSVCPFSVHSQKLGQGFLDCQ 240
Query 241 REKIYSSTPQNFVKRSIVLNGKYKEFDVWRSCYAYYFPRCKGFASKSSRERAYSYGIYDT 300
REKIYS TP+NF++ SIVLNGKYKEFDVWRSCY++++PRCKGF +KSSRERAYSY IYDT
Sbjct 241 REKIYSLTPENFIRSSIVLNGKYKEFDVWRSCYSFFYPRCKGFVTKSSRERAYSYSIYDT 300
Query 301 ARRLFSSSETTFSLAKEIAFYIKHFHFTDDTYLLDLFGHVSDQKSLLDLSNYFLDRDAMV 360
AR LF ++TTFSLAKEIA YI +FH +TYLLDL+G+ SDQ L +LS YF D D ++
Sbjct 301 ARLLFPDAKTTFSLAKEIAIYIYYFHNPKETYLLDLYGYCSDQSKLYELSQYFYDSDVLL 360
Query 361 RPVESDEFNRWVHRIYTELLVSKHFLYFVCTHTTLAERKSKQRMIEEFYSYLDYMHLTTF 420
S EF+R+VHRIYTELL+SKHFLYFVCTH TLAERKSKQR+IEEFYS LDYMHLT F
Sbjct 361 HSFNSGEFSRYVHRIYTELLISKHFLYFVCTHNTLAERKSKQRLIEEFYSRLDYMHLTKF 420
Query 421 FESQQEFYESDLIGDDDLCTDQWENSYYPYFYYNVHTNEP-FEKTPVYRLYASDVKKLFN 479
FE+QQ FYESDLIGDDDLCTD W+NSYYPYFY NV+T+ FEKTPVYRLY+SDVKKLFN
Sbjct 421 FEAQQLFYESDLIGDDDLCTDNWDNSYYPYFYNNVYTDTNLFEKTPVYRLYSSDVKKLFN 480
Query 480 DRIKHKKLNDANKIFID 496
DRIKHKKLNDANK+F +
Sbjct 481 DRIKHKKLNDANKVFFE 497
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 612 bits (1579), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 292/422 (69%), Positives = 350/422 (83%), Gaps = 1/422 (0%)
Query 77 LERPFGNDLVDKETGEILGPSDMKQEDIDRLLNKFYLFGDVPYLRKTDLQLFFKRLRYYV 136
+ERP+G+DLVD ETGE LG +D+ ++I+RL KF+LFG +PYLRK DLQLFFKR RYYV
Sbjct 1 MERPYGHDLVDVETGEYLGEADLSIKEIERLQEKFHLFGYLPYLRKFDLQLFFKRFRYYV 60
Query 137 SKQCPSEKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQVCSENISQAWTLGRIDCQISK 196
+K+ P EKVRYFA+GEYGPVHFRPHYHILLFLQSDEALQVCS+ +S+AW GR+DCQ+SK
Sbjct 61 AKRFPKEKVRYFAIGEYGPVHFRPHYHILLFLQSDEALQVCSKVVSEAWPFGRVDCQLSK 120
Query 197 GQCSSYVASYVNSSCTIPKVFKLSSVCPFNVHSQKLGQGFLDCQREKIYSSTPQNFVKRS 256
G+CSSYVA YVNSS +PKV L ++CPF VHSQKLGQGFL +R K+YS TP+ FVKRS
Sbjct 121 GKCSSYVAGYVNSSVLVPKVLTLPTLCPFCVHSQKLGQGFLQSERAKVYSLTPEQFVKRS 180
Query 257 IVLNGKYKEFDVWRSCYAYYFPRCKGFASKSSRERAYSYGIYDTARRLFSSSETTFSLAK 316
IV+NG+YKEFDVWRS YAY+FP+CKGFA KSSRERAYSYG+YDTARRLF S+ETTF+LAK
Sbjct 181 IVINGRYKEFDVWRSAYAYFFPKCKGFADKSSRERAYSYGLYDTARRLFPSAETTFALAK 240
Query 317 EIAFYIKHFHFTDDTYLLDLFGHVSDQKSLLDLSNYFLDRDAMVRPVESDEFNRWVHRIY 376
EI YI +FH DTY LD+FG VSDQ L S YF + + + ++S E R+VHR+Y
Sbjct 241 EIVGYIYYFHNKKDTYCLDIFGEVSDQSDLYQFSQYFFEPEIVNYSLDSIEMCRYVHRVY 300
Query 377 TELLVSKHFLYFVCTHTTLAERKSKQRMIEEFYSYLDYMHLTTFFESQQEFYESDLIGDD 436
TELL+SKHFLYFVC TL+E+K K ++IEEFYS LDYMHL TFFE+QQ FYESDL+GD
Sbjct 301 TELLLSKHFLYFVCDRPTLSEQKRKLKLIEEFYSRLDYMHLKTFFENQQLFYESDLVGDL 360
Query 437 DLCTDQWENSYYPYFYYNVH-TNEPFEKTPVYRLYASDVKKLFNDRIKHKKLNDANKIFI 495
DL +D WENSYYP+FY NV+ ++E ++KTPVYRLY + KLF+DRIKHKKLND NKIF+
Sbjct 361 DLMSDAWENSYYPFFYDNVYFSSEVYKKTPVYRLYDMQISKLFSDRIKHKKLNDLNKIFV 420
Query 496 DE 497
DE
Sbjct 421 DE 422
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 360 bits (925), Expect = 6e-115, Method: Compositional matrix adjust.
Identities = 215/527 (41%), Positives = 309/527 (59%), Gaps = 52/527 (10%)
Query 6 FCKCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYAN 65
F +CL PK++ NPY N+ ++VPCG C+AC +K SRY Q LE+ K +F TLTYAN
Sbjct 8 FIRCLEPKRVFNPYLNDWLLVPCGKCRACQCSKASRYKLQIQLEASQHKFCIFGTLTYAN 67
Query 66 RYIPRATFVDSLERPFGN----DLVDKETGEILGPSDMKQEDIDRLLNKFYLFGDVPYLR 121
YIPR + V ++ FG ++ DKETGE LG D D++ LL+K +LFGDVPYLR
Sbjct 68 TYIPRLSLVPYNDKTFGVVNGYEMCDKETGEYLGYLDSPSYDVESLLDKLHLFGDVPYLR 127
Query 122 KTDLQLFFKRLRYYVSKQCPSEKVRYFAVGEYGPVHFRPHYHILLFLQS----------- 170
K DLQLF KRLR +SK + KVRYFA+GEYGPVHFRPHYH LLF
Sbjct 128 KRDLQLFIKRLRKNLSKYSDA-KVRYFAMGEYGPVHFRPHYHFLLFFDEIKFTAPSGHTL 186
Query 171 -----------------DEALQVCSENISQAWTLGRIDCQISKGQCSSYVASYVNSSCTI 213
+ L V I +W GR+D Q SKG + YV+SYV+ S ++
Sbjct 187 GEFPDWAWYDSQNKCSRSDILSVVEYCIRSSWKFGRVDAQYSKGDAAQYVSSYVSGSGSL 246
Query 214 PKVFKLSSVCPFNVHSQKLGQGFLDCQREKIYSSTPQNFVKRSIVLNGKYKEFDVWRSCY 273
PKV+++SS PF++HS+ LGQGFL + EK+Y + ++FVKRS+ LNG K+F++WRSCY
Sbjct 247 PKVYQVSSARPFSLHSRFLGQGFLAHECEKVYETPVRDFVKRSVELNGSNKDFNLWRSCY 306
Query 274 AYYFPRCKGFASKSSRERAYSYGIYDTARRLFSSSETTFSLAKEIAFYIKHFHFTDDTYL 333
+ ++P+CKGF KSS ER Y+Y +YDTA+RLF + LA+E ++ + + +
Sbjct 307 SVFYPKCKGFTRKSSSERLYTYKLYDTAKRLFPYVSSVIELARETMIHLTFYVYGKQHTV 366
Query 334 LDLFGHVSDQKSLLDLSNYFLDRDAMVRPVESDE--FNRWVHRIYTELLVSKHFLYFVCT 391
+L D K L L+ + +V D+ ++ ++ IY ELL+S+HFL F C+
Sbjct 367 AEL---DYDIKRYLLYFRDSLNINEVVLQYGFDDVRIDKCIYLIYNELLLSRHFLEFCCS 423
Query 392 HTTLAERKSKQRMIEEFYSYLDYMHLTTFFESQQEFYESDLIGDDDLCTDQWENSYYPYF 451
+ + + IE FY LDY+ LT FF+SQ+ ++ D DD Y Y
Sbjct 424 GRS---QNFVFKRIEAFYKDLDYLQLTEFFKSQELYFSQDFCDSDD----------YVYM 470
Query 452 YYNVHTN-EPFEKTPVYRLYASDVKKLFNDRIKHKKLNDANKIFIDE 497
Y N + + ++++ Y + +++ +IKHK+LND N++F D+
Sbjct 471 YNNSSFSLDAYKQSMSYLSFEQQTFEIWRSKIKHKELNDLNQLFFDK 517
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 237 bits (604), Expect = 8e-68, Method: Compositional matrix adjust.
Identities = 168/513 (33%), Positives = 242/513 (47%), Gaps = 48/513 (9%)
Query 8 KCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYANRY 67
KC HP+ + N YT E + V CG C+AC+ + + +F C +E K+ +F TLTY+N Y
Sbjct 10 KCYHPRHVQNKYTGEVIQVGCGVCKACLKRRADKMSFLCAIEEQSHKYCMFATLTYSNDY 69
Query 68 IPRA--------------TFVDSLERPFGNDLVDKETGEILGPSDMKQEDIDRLLNKFYL 113
+PR ++ D L VD + D + L K L
Sbjct 70 VPRMYPEVDNELRLVRWYSYCDRLNEKGKLMTVDYDYWHKCPSLDTY---VLMLTAKCNL 126
Query 114 FGDVPYLRKTDLQLFFKRLRYYVSKQCPSEKVRYFAVGEYGPVHFRPHYHILLFLQSDEA 173
G + Y K D QLF KR+R +SK EK+RY+ V EYGP FR HYH+L F +
Sbjct 127 DGYLSYTSKRDAQLFLKRVRKNLSKY-SDEKIRYYIVSEYGPKTFRAHYHVLFFYDEVKT 185
Query 174 LQVCSENISQAWTLGRIDCQISKGQCSSYVASYVNSSCTIPKVFKLSSVCPFNVHSQKLG 233
+V S+ I QAW GR+DC +S+G+C+SYVA YVN + +P+ S PF+ HS +
Sbjct 186 QKVMSKVIRQAWQFGRVDCSLSRGKCNSYVARYVNCNYCLPRFLGDMSTKPFSCHSIRFA 245
Query 234 QGFLDCQREKIYSSTPQNFVKRSIVLNGKYKEFDVWRSCYAYYFPRCKGFASKSSRERAY 293
G Q+E+IY + +F+ +S +NG Y EF WR+ +FP+CKG++ KS E
Sbjct 246 LGIHQSQKEEIYKGSVDDFIYQSGEINGNYVEFMPWRNLSCTFFPKCKGYSRKSDSELWQ 305
Query 294 SYGIYDTARRLFSSS-ETTFSLAKEIAFYIKHFHFTDDTYLLDLFGHVSDQKSLLDLSNY 352
SY I R S T A+ I + F+ D+ G +L + +Y
Sbjct 306 SYNILREVRSAIGYSFNTIIDYARCILDLVVTAKFSCDSR-----GLPCSSPALNKVISY 360
Query 353 FLDRDAMVRPVESDEFNRW-VHRIYTELLVSKHFLYFVCTHTTLAERKSKQRMIEEFYSY 411
F + P SD + + I EL +S+HFL FVC + + ER K +I +F+
Sbjct 361 F-SQGIDTNPYYSDYLADYHTNSIARELYISRHFLTFVCDNDSYHERYRKFTLIRQFWQR 419
Query 412 LDYMHLTTFFESQQEFYESDLIGDDDLCTDQWENSYYPYFYYNVHTNEPFEKTPV----- 466
DY L + SQ E LI + Y ++Y N + F V
Sbjct 420 YDYALLVGMYTSQIE--NRHLISN------------YDWYYINKTPLDSFGNVDVSQLSK 465
Query 467 ---YRLYASDVKKLFNDRIKHKKLNDANKIFID 496
Y+ + + F IKHK NDAN FI+
Sbjct 466 ELFYKRFVIKSDENFEKSIKHKIQNDANGFFIN 498
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 223 bits (569), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 174/554 (31%), Positives = 275/554 (50%), Gaps = 82/554 (15%)
Query 6 FCKCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYAN 65
F CL P++I N YT E MVV C HC AC +N +Y+ CD ES AK T+F+TLT+ +
Sbjct 5 FVSCLEPQRIKNKYTGEEMVVACKHCVACEQLRNFKYSNLCDFESLTAKKTVFLTLTFDD 64
Query 66 RYIPRATFVDSLERPFGND---LVDKETGEILGPSDMKQEDID----RLLNKFYLFGDVP 118
+++P+ F G+D + D +TGE LG + M + ++ R+ + G P
Sbjct 65 KFVPQFRFYK-----VGDDEYIMRDADTGEYLGRTLMTPQLMNEYQKRVNYRINYKGRFP 119
Query 119 YLRKTDLQLFFKRLRYYVSKQCPSEKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQVCS 178
YL K +LQLF KRLR Y+ K +K+R+FA GEYGP+ FRPH+HILLF+ D +L + S
Sbjct 120 YLSKRELQLFMKRLRKYLDKY-EGQKIRFFATGEYGPLSFRPHFHILLFV-DDPSLFLPS 177
Query 179 EN------------------------------ISQAWTLGRIDCQ-ISKGQCSSYVASYV 207
+ I ++W G ID Q + +G CSSYVA YV
Sbjct 178 VHTLGEYPYPYWSKYQKAHCGKGTLLSKLEYYIRESWPFGGIDAQSVEQGSCSSYVAGYV 237
Query 208 NSSCTIPKVFKLSSVCPFNVHSQKLGQGFLDCQREKIYSSTPQNFVKRSIVLNGKYKEFD 267
NSS +P K+ +V F+ HS+ LG+ + + FV+RS G+Y F
Sbjct 238 NSSVPLPSCLKVDAVKSFSQHSRFLGRKIFGTELIPLLKLKFTEFVQRSFFCRGRYDNFR 297
Query 268 VWRSCYAYYFPRCKGFASKSSRERAYSYGIYDTARRLFSSSETTFSLAKEIAFYIKHFHF 327
+P+CKGFA S +R Y I+ R F+S + +A+ + + F+
Sbjct 298 TPSEMLHSVYPQCKGFALLSHEQRFRVYTIWSRLRYYFNSDKKA-DVARSL---VTSFYS 353
Query 328 TDDTYLLDLFGHVSDQKSLL------DLSNYFLDRDAMVRPVESDEFNRWVHR-IYTELL 380
DT +L + V + L+ +L+ +DR + D+ N + + +Y+ LL
Sbjct 354 WLDTGILRVPERVREDFLLIYTELSQNLNYKRIDRFDYDKFRHDDDLNNQLFQCVYSILL 413
Query 381 VSKHFLY-------FVCTHTTLA----ERKSKQRMIEEFYSYLDYMHLTTFFESQQEFYE 429
S F + ++C+ + + + R +E F+ +Y++L ++ Q+ +++
Sbjct 414 CSSVFEHNAKLWKSYLCSLSLMWCDFDRNELFLRKVERFWKNYEYLNLVDWYRKQEIYFD 473
Query 430 ------SDLI-GDDDLCTDQWENSYYPYFYYNV-HTNEPFEK-TPVYRLYASDVKKLFND 480
SD + G + L D YFY NV + E F+K T Y+ Y+++V+ +
Sbjct 474 KSYSRKSDFLDGKERLSGD------IKYFYNNVPYDVEQFKKRTFAYKAYSANVRFMARQ 527
Query 481 RIKHKKLNDANKIF 494
R+KHK+ ND N IF
Sbjct 528 RMKHKEQNDKNMIF 541
>gi|494610270|ref|WP_007368516.1| hypothetical protein [Prevotella multiformis]
gi|324988542|gb|EGC20505.1| hypothetical protein HMPREF9141_0984 [Prevotella multiformis
DSM 16608]
Length=479
Score = 125 bits (313), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 90/299 (30%), Positives = 138/299 (46%), Gaps = 17/299 (6%)
Query 9 CLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYANRYI 68
CL PK+I N Y +E++ VPC C C + S ++ + + E + +LF+TLTY N +I
Sbjct 10 CLSPKRIYNKYIDETLYVPCRKCFRCRDSYASDWSRRIENECREHRFSLFVTLTYDNEHI 69
Query 69 P--RATFVDSLERP--FGNDLVDKETGEILGPSDMKQEDIDRLLNKFYLFGDVPYLRKTD 124
P + +D P F N L E+G+ L S + ++ ++ Y K D
Sbjct 70 PLFQPLVMDDGSHPVWFSNRL--SESGKFLSDSVCRSLPPQKMEDEVCF----AYPCKKD 123
Query 125 LQLFFKRLRYYVSKQCPSEK-----VRYFAVGEYGPVHFRPHYHILLFLQSDEALQVCSE 179
+Q +FKRLR V Q K +RYF EYGP FRPHYH +L+ S+E +
Sbjct 124 VQDWFKRLRSAVDYQLNKNKSNEFRIRYFICSEYGPRTFRPHYHAILWYDSEELQRNIGR 183
Query 180 NISQAWTLGRIDCQISKGQCSSYVASYVNSSCTIPKVFKLSSVCPFNVHSQKLGQGFLDC 239
I + W G + S YVA YVN +P + F++ S+ G+
Sbjct 184 LIRETWKNGNSVFSLVNNSASQYVAKYVNGDTRLPPFLRTEFTSTFHLASKHPYIGYCKA 243
Query 240 QREKIYSSTPQNFVKRSIVL--NGKYKEFDVWRSCYAYYFPRCKGFASKSSRERAYSYG 296
E + S+ +S++ NG+++ RS P+C+G+ S S ER Y
Sbjct 244 DEEALRSNVLDGTYGQSVLNRDNGQFEFVPTPRSLENRLLPKCRGYRSLSHSERIRVYA 302
>gi|565841285|ref|WP_023924566.1| hypothetical protein [Prevotella nigrescens]
gi|564729906|gb|ETD29850.1| hypothetical protein HMPREF1173_00032 [Prevotella nigrescens
CC14M]
Length=484
Score = 123 bits (308), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 82/251 (33%), Positives = 126/251 (50%), Gaps = 45/251 (18%)
Query 9 CLHPKKIVNPYTNESMVVPCGHCQACMLAKNS----RYAFQCDLESYVAKHTLFITLTYA 64
C HPK+I+NPYT+E + V C C+ C+ K S R A +C L +Y A F+TLTY
Sbjct 9 CEHPKRIINPYTHERVWVACRRCKCCLNKKTSAWSGRVANECKLHAYSA----FVTLTYD 64
Query 65 NRYIPRATFVDSLERPFGNDLVDKETGEILGPSDMKQEDIDRLLNKFYLFGD-------- 116
N ++P L +P + E GE++ S+ RL ++ + G+
Sbjct 65 NEHLP-------LYQPECMN----ERGEMVWTSN-------RLCDEKVIVGNYDFIKVSN 106
Query 117 -----VPYLRKTDLQLFFKRLR----YYVSKQ--CPSEKVRYFAVGEYGPVHFRPHYHIL 165
V Y K+D+ FFKRLR YY K +EK+RYF EYGP RPHYH +
Sbjct 107 SDVQAVAYCCKSDIVKFFKRLRSKLSYYFKKHHIITNEKIRYFVCSEYGPKTLRPHYHAI 166
Query 166 LFLQSDEALQVCSENISQAWTLGRIDCQISKGQCSSYVASYVNSSCTIPKVFKLSSVCPF 225
++ S+E +V + +S +W+ G D + YVA YV+ + +P++ + + F
Sbjct 167 IWFDSEEVARVIEKMLSSSWSNGFTDFEYVNSTAPQYVAKYVSGNSVLPEILQHDACRTF 226
Query 226 NVHSQKLGQGF 236
++ SQ G+
Sbjct 227 HLQSQAPSVGY 237
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 114 bits (285), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 111/375 (30%), Positives = 164/375 (44%), Gaps = 57/375 (15%)
Query 1 MIKNSFCKCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFIT 60
MI+ SF KC P + +P VPCG C AC K S + + LE Y +K+ F+T
Sbjct 1 MIQ-SFVKCFSPLVLRDP-RGYPYQVPCGKCIACHNNKRSSLSLKLRLEEYTSKYCYFLT 58
Query 61 LTYANRYIP---------RATFV-----------DSLERPFGNDLVDKETGEILGPSDMK 100
LTY + +P FV DS F +DL + + + + D
Sbjct 59 LTYDDDNLPLFSVGLDTCATEFVRIYPYSERLRNDSFISDFCSDLHNFD-NDFVDKMDYY 117
Query 101 QEDIDRLLNKF-----YLFGDVPYLRKTDLQLFFKRLRYYVSKQCPSEKVRYFAVGEYGP 155
+ + +K+ Y G L D+QLF KRLR ++ K EK+R++ +GEYG
Sbjct 118 SDYVINYESKYHKSCVYGHGLYALLYYRDIQLFLKRLRKHIYKY-YGEKIRFYIIGEYGT 176
Query 156 VHFRPHYHILLFLQSDEALQV---------------CSENISQAWTLGRIDCQISKGQCS 200
RPH+H LLF S Q C + W G D + + G+
Sbjct 177 KSLRPHWHCLLFFNSSSLSQAFEDCVNVGTTSRPCSCPRFLRPFWQFGICDSKRTNGEAY 236
Query 201 SYVASYVNSSCTIPKVFKLSSVCPFNVHSQKLGQGFLDCQREKIYSSTPQ---NFVKRSI 257
+YV+SYVN S PK+ L S HS +LGQ + I S+ + +F +R
Sbjct 237 NYVSSYVNQSANFPKLLVLLSNQK-AYHSIQLGQIL---SEQSIVSAIQKGDFSFFERQF 292
Query 258 VLN--GKYKEFDVWRSCYAYYFPRCKGFASKSSRERAYS-YGIYDTARRLFSSSETTFSL 314
L+ G + VWRS Y+ +FP+ +S+ + E+ Y Y+T R LF +
Sbjct 293 YLDTFGAANSYSVWRSYYSRFFPKFTC-SSQLTYEQTYRVLTCYETLRDLFDTDSVGVIC 351
Query 315 AKEIAFYIKHFHFTD 329
+ FY HF + D
Sbjct 352 RR--LFYHYHFGYPD 364
>gi|647452984|ref|WP_025792805.1| hypothetical protein [Prevotella histicola]
Length=480
Score = 113 bits (282), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 89/298 (30%), Positives = 130/298 (44%), Gaps = 13/298 (4%)
Query 9 CLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYANRYI 68
CL P +I N Y E + C C C + S +A + D E +++LF+TLTY N ++
Sbjct 12 CLRPHRIYNRYIGEFLYTNCRKCVRCRSSYASSWANRIDSECSFHRYSLFLTLTYDNDHL 71
Query 69 PRATFVDSLERPFGNDLVDKETGEILGPSDMKQEDIDRLLNKFYLFGDV--PYLRKTDLQ 126
P + +L+ D+ G G DI R + + V Y K D+Q
Sbjct 72 PYYAPLFNLDGS-RTDVWCSNRGCDNG--KFVSSDIARPIPPVGMEDTVCFAYPCKKDVQ 128
Query 127 LFFKRLR----YYVSKQCPSEKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQVCSENIS 182
FFKRLR Y + + ++RYF EYGP FRPHYH +L+ S+ + I
Sbjct 129 DFFKRLRSKIDYKLKPRGNEYRIRYFICSEYGPNTFRPHYHAILWYDSEILHNELNVLIR 188
Query 183 QAWTLGRIDCQISKGQCSSYVASYVNSSCTIPKVFKLSSVCPFNVHSQKLGQGFLDCQRE 242
+ W G D + S YVA YVN C +P + F++ S+ G+ E
Sbjct 189 ETWKNGNTDFSLVNSSASQYVAKYVNGDCDLPSFLRTEFTSTFHLASKHPCIGYGKDDEE 248
Query 243 KIYSSTPQNFVKRSIVLNGKYKEFDVW---RSCYAYYFPRCKGFASKSSRERAYSYGI 297
+Y + R+ LN EF+ RS P+CKG+ S ER Y +
Sbjct 249 ALYENVINGTYGRN-CLNKSTNEFEFVCPPRSLENRILPKCKGYRRISHSERVRIYAL 305
>gi|496521300|ref|WP_009229583.1| hypothetical protein [Prevotella sp. oral taxon 317]
gi|288330571|gb|EFC69155.1| hypothetical protein HMPREF0670_00478 [Prevotella sp. oral taxon
317 str. F0108]
Length=569
Score = 110 bits (275), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 96/344 (28%), Positives = 149/344 (43%), Gaps = 61/344 (18%)
Query 6 FCKCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYAN 65
F CL P + N +T + M VPCG C+AC+ A S+ + + E K+++ TLTY N
Sbjct 8 FGNCLCPVHVHNRWTRDEMFVPCGRCEACVNAAASKQSKRVRNEIMQHKYSVMFTLTYNN 67
Query 66 RYIPR-ATFVDSLE----RPFGN----------DLVDKETGEILGPSDMKQEDIDRLLNK 110
+IPR F+D+ + RP G + DK TG+ D+D L K
Sbjct 68 EFIPRWERFLDNNDCPQLRPIGRCAELFPSCPLNYFDKVTGKW-------SIDLDTFLPK 120
Query 111 F------YLFGDVPYLRKTDLQLFFKRLRYYVSK---QCPSEKVRYFAVGEYGPVHFRPH 161
+F K D+Q F KRLR+ +SK + S K+RY+ EYGP RPH
Sbjct 121 IENDEHTEVFASCC---KKDIQNFLKRLRFNISKLYGKAESRKIRYYVASEYGPTTLRPH 177
Query 162 YHILLFLQSDEALQVCSENISQAWTLGR--------------IDCQISK-------GQCS 200
YH ++F L S I ++W R D +++ +
Sbjct 178 YHGIIFFDDASLLSEISSLIVRSWGFQRRVGGKRNSFIFQPFADISLTQQYVKLCDQNTA 237
Query 201 SYVASYVNSSCTIPKVFKLSSVCPFNVHSQKLGQGFLDCQREKIYSSTPQNF--VKRSIV 258
YVA YV+ + +P+V S PF++ S+ G ++ + V R +
Sbjct 238 YYVAEYVSGNLGLPQVLAYKSTLPFHLCSKSPVIGCFKADYCEVLGRVHRGAYRVGREVF 297
Query 259 --LNGKYKEFDVW--RSCYAYYFPRCKGFASKSSRERAYSYGIY 298
+G++ +D+ R + F +C GF+S S E+ Y Y
Sbjct 298 DEKSGQFMHYDIPLDRDLCSSLFRKCLGFSSLSFNEKLLRYSFY 341
Lambda K H a alpha
0.324 0.138 0.429 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3489190903935